Figures & data
Figure 1 Synthesis of phenylthiazole acids.
Abbreviations: DMF, dimethylformamide; rt, room temperature.
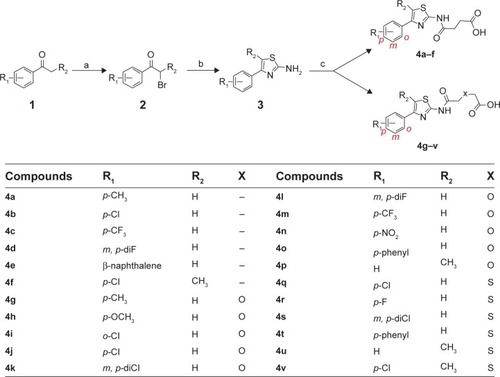
Figure 2 Fluorescence polarization (FP)-based PPARγ ligand screening assay of phenylthiazoles.
Abbreviations: PPARγ, peroxisome proliferator-activated receptor gamma; RSG, rosiglitazone.
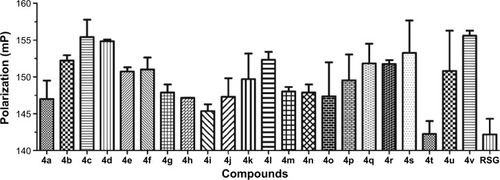
Table 1 EC50s of phenyl thiazole acids in FP-based PPARγ ligand screening assay
Table 2 The molecular properties and binding energy of selected phenylthiazole acids
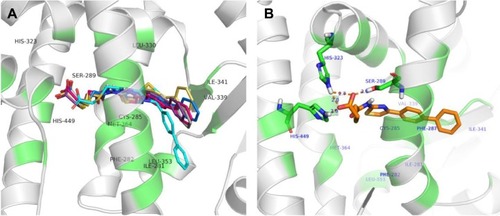
Figure 3 The binding modes of agonists with PPARγ. Orange and cyans colors represented RSG and other agonists, respectively.
Abbreviations: PPARγ, peroxisome proliferator-activated receptor gamma; RSG, rosiglitazone.
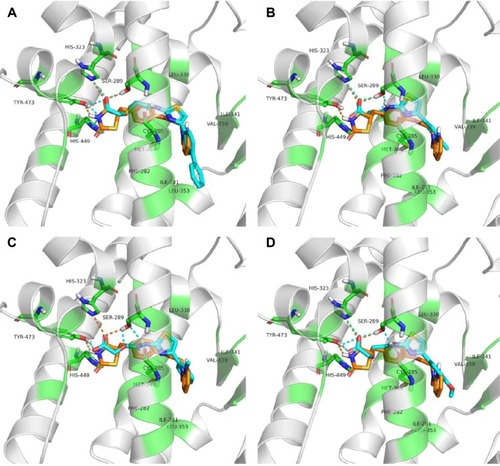
Figure 4 The RMSD of ligands from the starting structure during 25 ns MD simulations.
Abbreviations: RMSD, root mean square deviation; MD, molecular dynamics; PPARγ, peroxisome proliferator-activated receptor gamma; RSG, rosiglitazone.
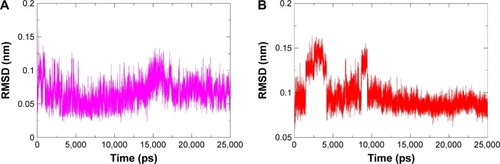
Figure 5 The binding modes of 4t.
Abbreviations: PPARγ, peroxisome proliferator-activated receptor gamma; MD, molecular dynamics.