Figures & data
Table 1 Pathogenic alleles that cause α1-antitrypsin deficiency
Figure 1 (A) Inhibition of neutrophil elastase by α1-antitrypsin. After docking (left) the neutrophil elastase (grey) is inactivated by movement from the upper to the lower pole of the protein (right). This is associated with insertion of the reactive loop (red) as an extra strand into β-sheet A (green). Reproduced from Lomas et alCitation136 with permission. (B) The structure of α1-antitrypsin is centered on β-sheet A (green) and the mobile reactive center loop (red). Polymer formation results from the Z variant of α1-antitrypsin (Glu342 Lys at P17; arrowed) or mutations in the shutter domain (blue circle) that open β-sheet A to favor partial loop insertion (step 1) and the formation of an unstable intermediate (M*). The patent β-sheet A can either accept the loop of another molecule (step 2) to form a dimer (D), which then extends into polymers (P). A small proportion of the unstable serpin molecules can accept their own loop (step 3) to form an inactive, thermostable, latent conformation (L). The individual molecules of α1-antitrypsin within the polymer are colored red, yellow, and blue. Reproduced from Gooptu et alCitation29 with permission.
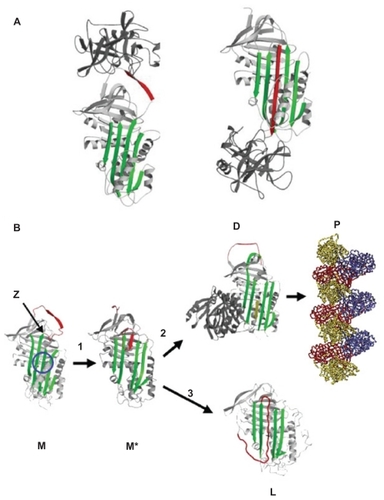
Figure 2 (A) Electron microscopy (×20,000) of a hepatocyte from a Z homozygote showing a massive inclusion (arrowed) in the endoplasmic reticulum. Reproduced from Lomas et alCitation18 with permission. (B) The intra-hepatic polymers of mutant Z α1-antitrypsin have the appearance of beads on a string on electron microscopy. Reproduced from Lomas et alCitation137 with permission.
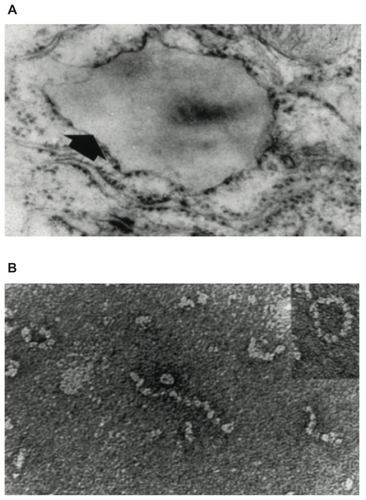
Table 2 Studies of α1-antitrypsin replacement therapy