Figures & data
Table 1 Nucleotide sequences of primers used for PCR
Figure 1 Effects of quercetin on the cell viability of ARPE-19 cells stimulated by TGF-β1.
Abbreviations: CCK-8, cell counting kit-8; TGF-β, transforming growth factor-β.
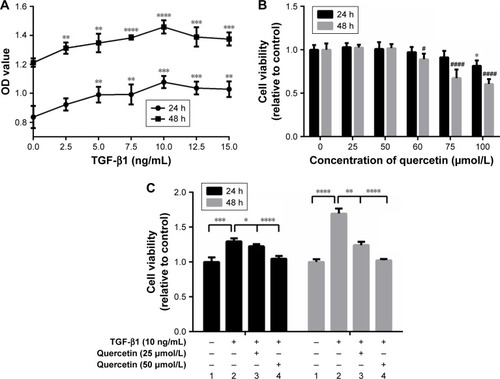
Figure 2 Effects of quercetin on wound closure in TGF-β1-treated RPE cells.
Abbreviations: que, quercetin; RPE, retinal pigment epithelial; TGF-β, transforming growth factor-β.
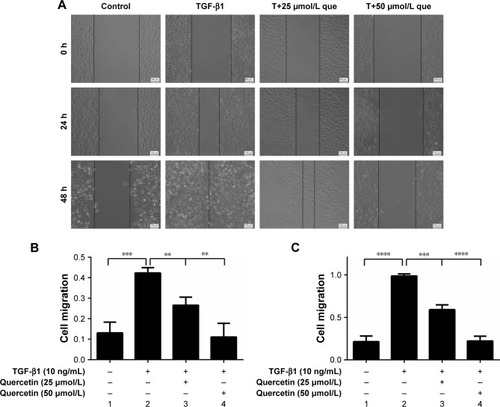
Figure 3 Effects of quercetin on cell migration in TGF-β1-treated RPE cells.
Abbreviations: que, quercetin; RPE, retinal pigment epithelial; TGF-β1, transforming growth factor-β1.
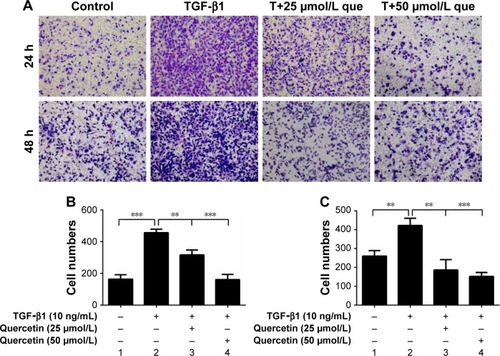
Figure 4 Effect of quercetin on collagen I secretion in TGF-β1-treated RPE cells.
Abbreviations: RPE, retinal pigment epithelial; TGF-β1, transforming growth factor-β1.
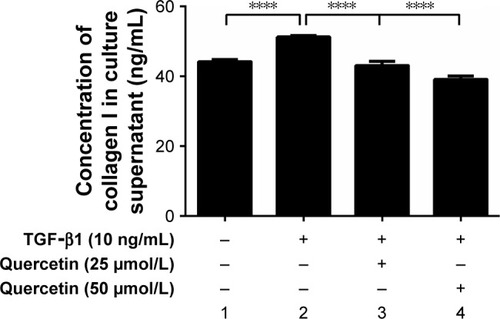
Figure 5 Quercetin suppressed TGF-β1-induced MMP expression in RPE cells.
Abbreviations: MMP-2, matrix metalloproteinase-2; MMP-9, matrix metalloproteinase-9; que, quercetin; RPE, retinal pigment epithelial; TGF-β1, transforming growth factor-β1.
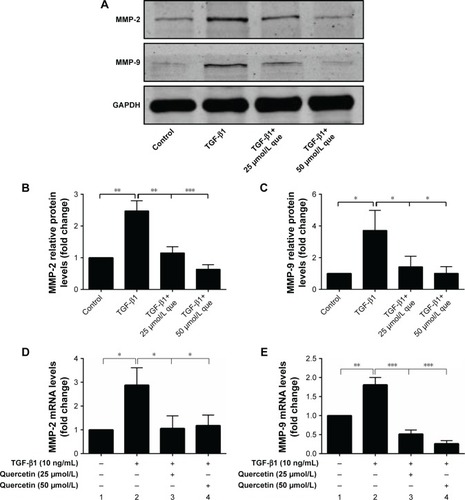
Figure 6 Quercetin suppressed TGF-β1-induced EMT in RPE cells.
Abbreviations: α-SMA, α-smooth muscle actin; EMT, epithelial–mesenchymal transition; que, quercetin; RPE, retinal pigment epithelial; ZO-1, zonula occludens-1.
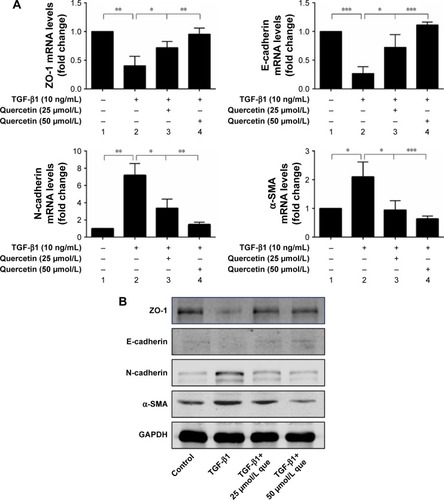
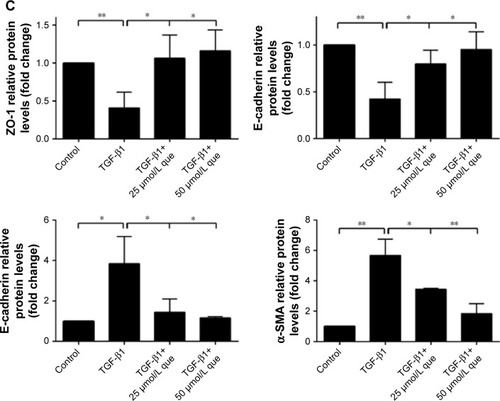
Figure 7 Immunofluorescence analysis of EMT-related proteins in ARPE-19 cells.
Abbreviations: α-SMA, α-smooth muscle actin; Ecad, E-cadherin; EMT, epithelial–mesenchymal transition; Ncad, N-cadherin; que, quercetin; RPE, retinal pigment epithelial; TGF-β1, transforming growth factor-β1; ZO-1, zonula occludens-1.
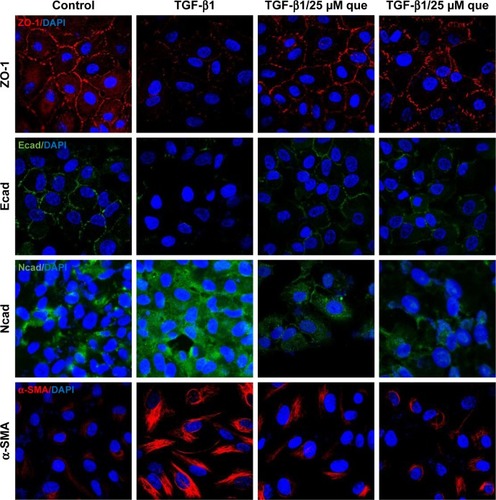
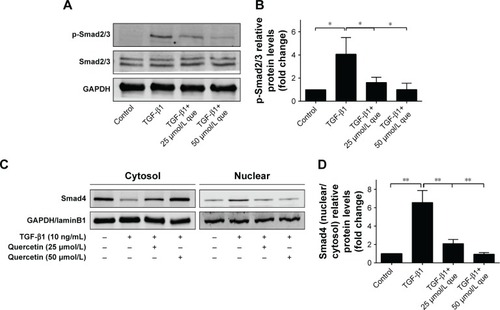
Figure 8 Quercetin attenuates TGF-β1-induced Smad2/3 phosphorylation and nuclear translocation of Smad4 in RPE cells.
Abbreviations: que, quercetin; RPE, retinal pigment epithelial; TGF-β1, transforming growth factor-β1.