Figures & data
Table 1 General description of the native input vector
Figure 4 Results of sensitivity analysis for the most important 28 inputs, with relative importance computed in the context of the native dataset.
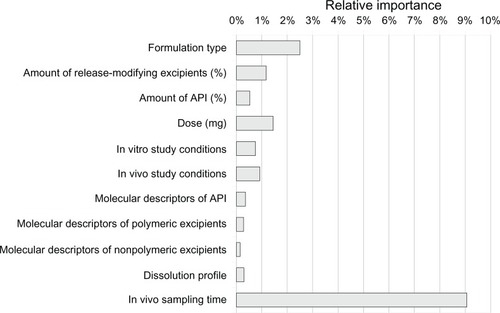
Table 2 Input vector reduced to 28 governing variables
Table 3 Architecture of ANNs selected for expert committee and their generalization errors
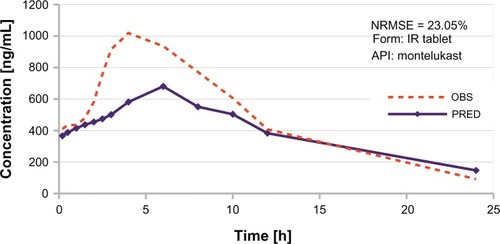
Figure 5 Examples of in vivo profile predictions.
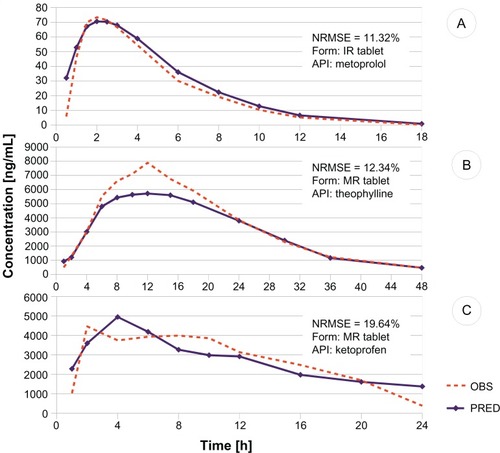
Figure 6 Example of in vivo profile prediction for the new test dataset introduced after the model development phase.