Figures & data
Table 1. Protein content and elastase inhibitor content of Macadamia nut, Brazil nut, soybean, and marama bean.
Figure 1. Elution profile of elastase and trypsin inhibitory activity from marama bean extracts using an elastase affinity column. The column wash included fractions 1–50 and the citrate elution fractions 51–77. Protein quantity measured at 280 nm is illustrated by the dotted line, trypsin inhibitory activity by squares, and elastase inhibition by open circles. Each fraction was 2 mL. Inhibitory activity was expressed as a percent of the control with no added inhibitor.
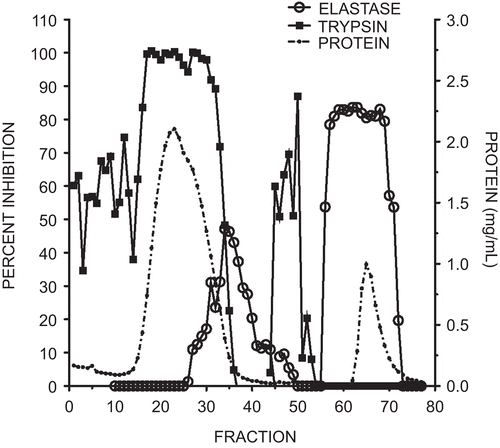
Figure 2. Elution profile of elastase and trypsin inhibitory activity from soybean trypsin inhibitor using an elastase affinity column. The column wash included fractions 1–50 and the citrate elution included fractions 51–77. Protein measured at 280 nm is illustrated by the dotted line, trypsin inhibitory activity by squares, and elastase inhibition by open circles. Each fraction was 2 mL. Inhibitory activity was expressed as a percent of the control with no added inhibitor.
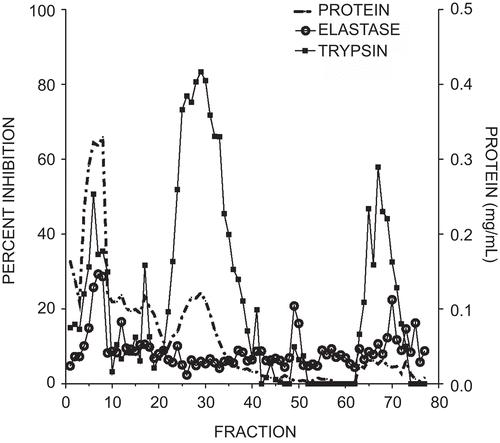
Figure 3. SDS PAGE of purified elastase inhibitor compared with soybean trypsin inhibitor. Lane 1, standard proteins with molecular mass shown; lane 2, two intensely-staining bands of apparent 17.8 kDa and 20 kDa in a reduced sample of the marama bean extract purified through the affinity column; lane 3, separation of reduced soybean trypsin inhibitor indicating major bands at 23.1 kDa and 24.0 kDa.
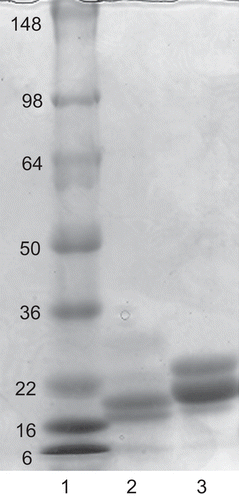
Figure 4. Tandem mass spectrum of a 1893.1-Da tryptic peptide from the marama bean elastase inhibitor; assigned b-series (N-termal) and y-series (C-teminal) are indicated above the corresponding peaks and on the peptide sequence.
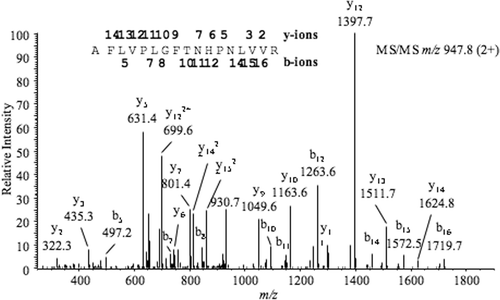
Figure 5. Sequence alignment of gi145579433 (Bbki, a Kunitz-type Kallikrein inhibitor, chain A) with peptides from the marama bean elastase inhibitor. Initial BLAST search indicated homology with gi145579433. A search of the peptides against Pfam family PF00197 using the Sequence Alignment Modeling system (SAMCitation31,Citation32) indicated that the peptides shown are significant matches.

Table 2. De novo sequences obtained from HPLC-ESI-MS/MS analysis of the elastase-specific inhibitor in marama bean.