Figures & data
Figure 1. The initial structure of the tetrameric M2 protein complexed with four rimantadines (RMTs) bound outside the channel in the pre-equilibrated lipid bilayer-water pieces where L represents the distance along a channel axis starting from the extracellular site. The structure of the His37 tetrad has been coloured orange. Close view of the RMT-Asp44-Trp41 hydrogen bond network with definitions of the d1–d4 distances are also shown.
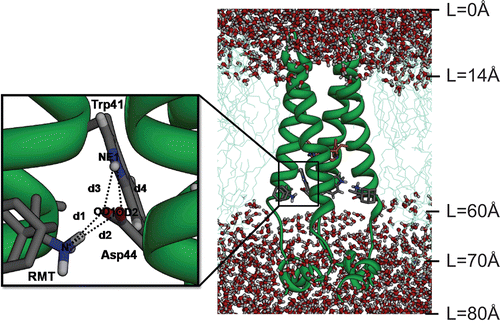
Figure 2. The root mean square displacements (RMSD) for the M2-RMT complexes in the 0H, 1H and 3H states.
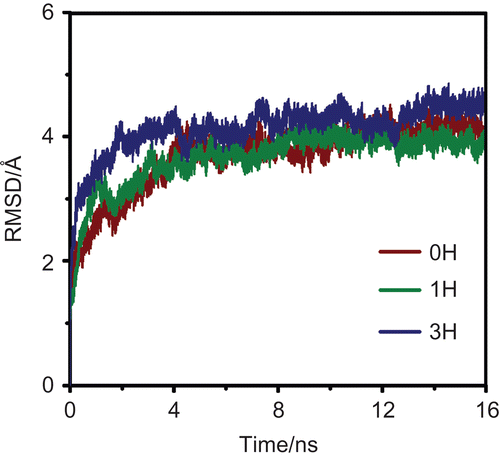
Table 1. Percentage of hydrogen bonds in the three (0H, 1H and 3H) protonation states between (i) the ammonium group of RMT and M2 residues and (ii) the Asp44 and Trp41 gating residues.
Figure 3. The distributions of the d1–d4 distances, as defined in , for the three simulated systems: 0H, 1H and 3H, at the subunits I–IV of the M2 channel.
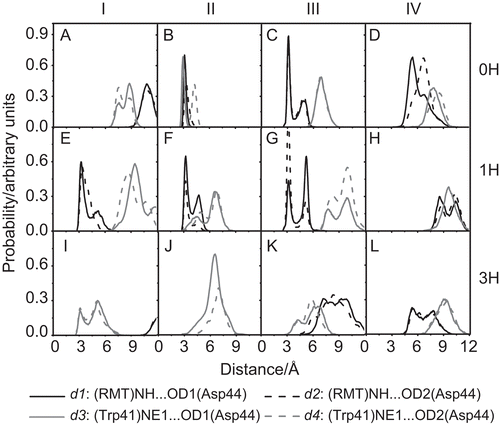
Figure 4. The water densities and the distribution patterns of the His37 selectivity and Trp41 gating residue positions in the free M2 protein and M2-RMTs complex.
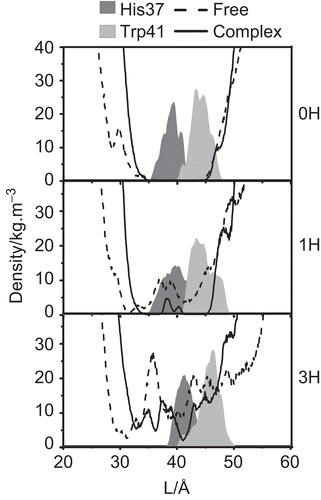
Figure 5. The rotational angle defined by CA, CB, CG and CD2 of the M2 Trp41 gate when M2 is complexed with the four RMTs outside the pore at different protonation states (0H, 1H and 3H). I–IV denote the four M2 subunits. Here, the vertical grey solid line (at 75°) represents the tortional angle of the closed conformation of the Trp41 indole ring obtained from NMR structure.
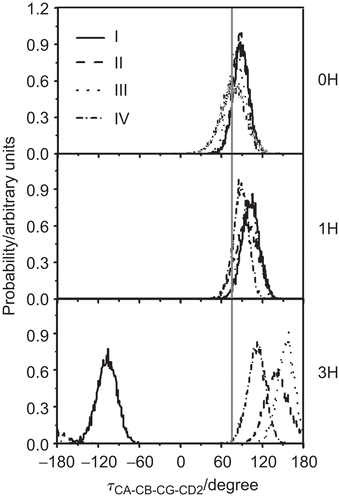
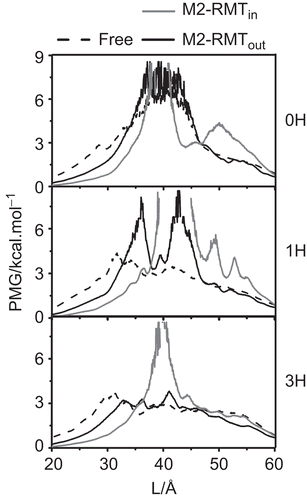
Figure 6. The potential of mean force (PMF) of the excess proton along the pore channel axis (L) starting from the N-terminal site for the three protonation states, 0H, 1H and 3H of the free M2 (dashed line) and RMT complexes where the four RMTs were located outside (black solid line, M2-RMTout) and inside the ion channel (grey solid line, M2-RMTin).