Figures & data
Table 1. Uni-Column statistics for training and test set of compounds.
Table 2. Statistical results of 2D-QSAR equation generated by MLR method and 3D-QSAR model generated by SA kNN–MFA for benzyl urea derivatives.
Table 3. Structure of training and test sets of compounds along with observed and predicted activity.
Figure 3. Comparison of observed activity vs predicted activity for training set (A) and test set (B) of compounds.
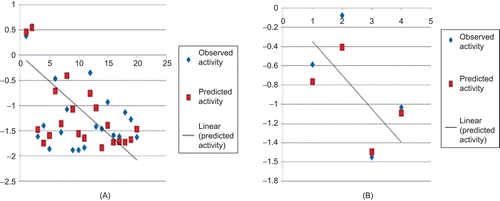
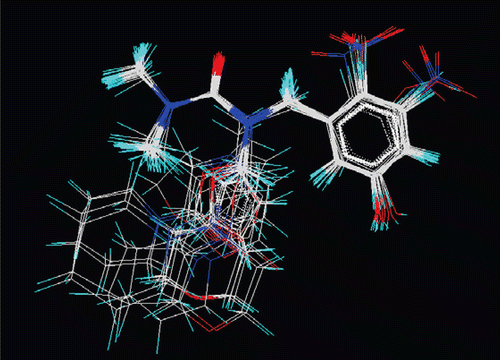
Figure 4. Results of QSAR studies. (A) Contributions of 2D descriptors for biological activity developed using MLR equation (2D-QSAR). (B) Contributions of electrostatic and steric 3D data points towards biological activity developed using kNN–MFA method (3D-QSAR). (C) Pharmacophore requirement around benzyl urea.
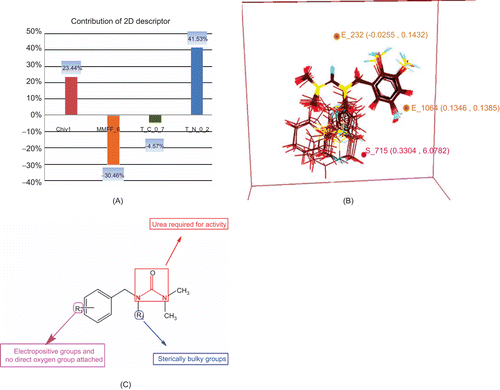
Table 4. Structure of designed NCEs along with predicted activity obtained by MLR equation generated by 2D-QSAR.
Table 5. Prediction of ADMET properties by using discovery studio, accelrys.
Scheme 1. Synthetic route for benzyl urea derivatives. Reagents & condition: (i) ethanol, reflux, 1–2 h (ii) NaBH4, 50°C, 2–3 h (iii) dry CH2Cl2, 70°C, 4 h (iv) dry CH2Cl2, dry pyridine, 60°C, 5–6 h.
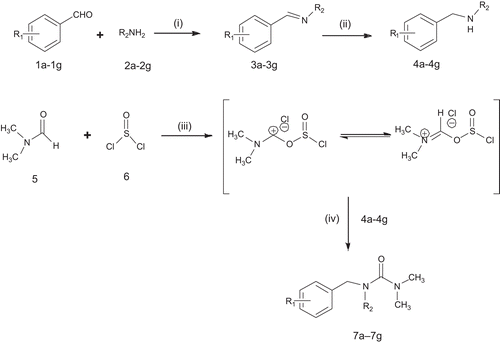
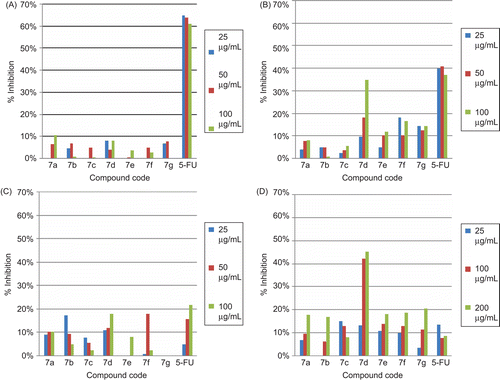
Table 6. Anti-proliferative activity of compounds 7a–7g in cancer cell lines.