Figures & data
Table 1. The structures of the training and test set molecules.
Table 2. The actual pIC50s, predicted pIC50s (Pred.) and their residuals (Res.) of the training and test set molecules.
Figure 3. Molecular Computer Aided Design (MOLCAD) Robbin surface of the adenosine triphosphate-binding site of VEGF-R2 (PDB code: 1Y6A) within compound 29. Key residues and hydrogen bonds were labelled. The α-helices were shown as helices or cylinders, while β-sheets were shown as arrows and the loop regions as tubes.
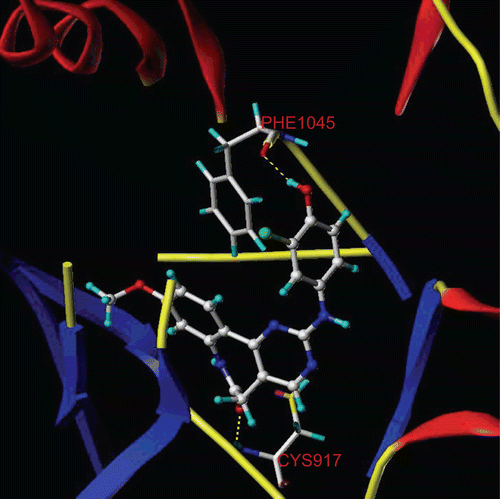
Figure 4. The Molecular Computer Aided Design (MOLCAD) Robbin and multi-channel surfaces structure displayed with cavity depth (CD) potential of the adenosine triphosphate pocket of VEGF-R2 (PDB code: 1Y6A) within compound 29. The cavity depth colour ramp ranges from blue (low depth values = outside of the pocket) to light red (high depth values = cavities deep inside the pocket).
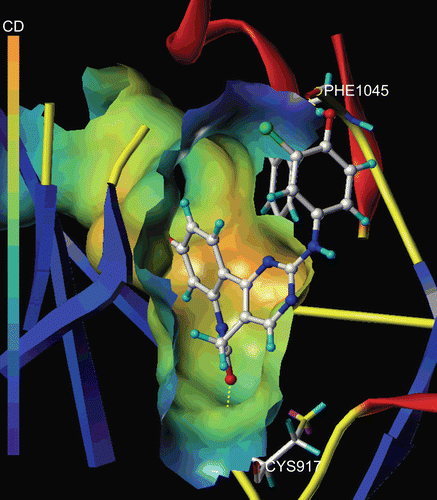
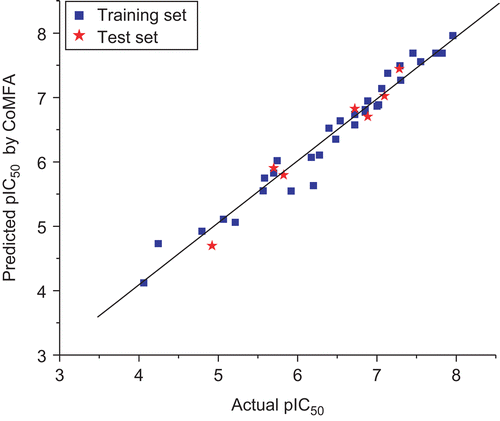
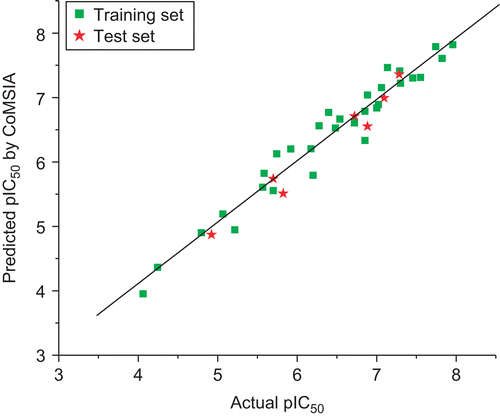
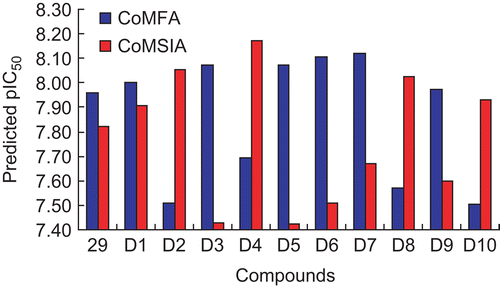
Table 3. Results of CoMFA and CoMSIA models.
Figure 7. Standard coefficient contour maps of CoMFA analysis with 2 Å grid spacing in combination with compound 29. (A) Steric fields: green contours (80% contribution) indicate regions where bulky groups increase activity, while yellow contours (20% contribution) indicate regions where bulky groups decrease activity, and (B) electrostatic fields: blue contours (80% contribution) indicate regions where electron-donating groups increase activity, while red contours (20% contribution) indicate regions where electron-withdrawing groups increase activity.
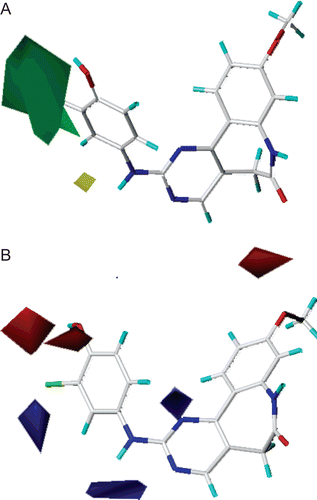
Figure 8. Standard coefficient contour maps of CoMSIA analysis with 2 Å grid spacing in combination with compound 29. (A) Steric contour map. Green and yellow contours refer to sterically favoured and unfavoured regions. (B) Electrostatic contour map. Blue and red contours refer to regions where electron-donating and electron-withdrawing groups are favoured. (C) Hydrophobic contour map. White and yellow contours refer to regions where hydrophilic and hydrophobic substituents are favoured. (D) Hydrogen bond donor and acceptor contour map. The cyan and purple contours indicate favourable and unfavourable hydrogen bond donor groups. The magenta and red contours demonstrated favourable and unfavourable hydrogen bond acceptor groups.
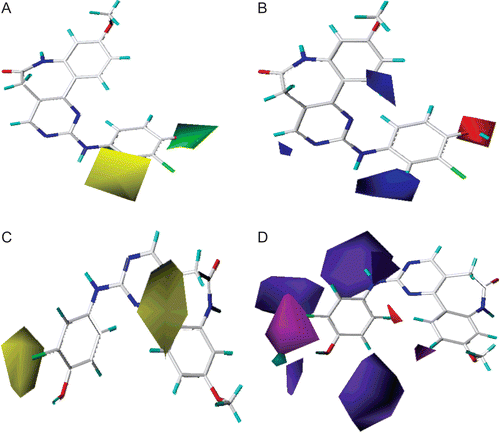
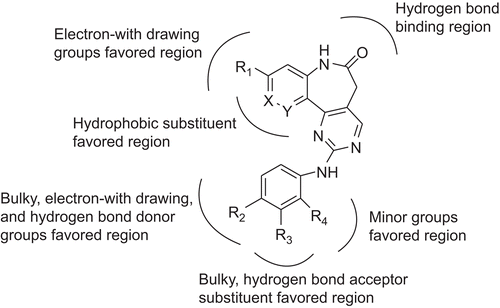
Table 4. Chemical structures of newly designed inhibitors and their predicted pIC50 values.