Figures & data
Table 1. Activity and binding properties of malbrancheamides analogues on the Ca2+-CaM complex.
Figure 2. Fluorescence spectra and titration curves of Ca2+-hCaM-M124C-mBBr in the presence of 1 (A), 2 (B), 3 (C), and 4 (D). Buffer was 100 mM of potassium acetate (pH 5.1) at 37°C and 1 mM CaCl2. Samples were excited at 381 nm, and emission spectra recorded for light scattering effects from 400 to 550 nm. The absolute changes of maximal fluorescence emission were plotted against the ration alkaloids/protein total and fitted to the binding equation model to obtain the Kd and stoichiometric ration.
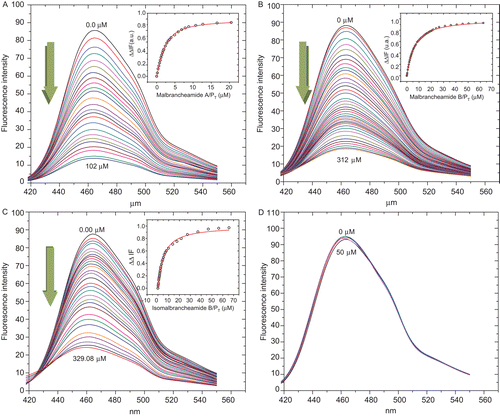
Figure 3. Far-UV circular dichroism of hCaM M124C-mBBr. in presence of 1 mM CaCl2. Buffer (−); Ca2+-hCaM M124C-mBBr (––); Ca2+-hCaM M124C-mBBr−1 (...); and Ca2+-hCaM M124C-mBBr−4 (–.–). The estimation of secondary structures from circular dichroism spectral in presence of all ligands was determined using K2D2 program.
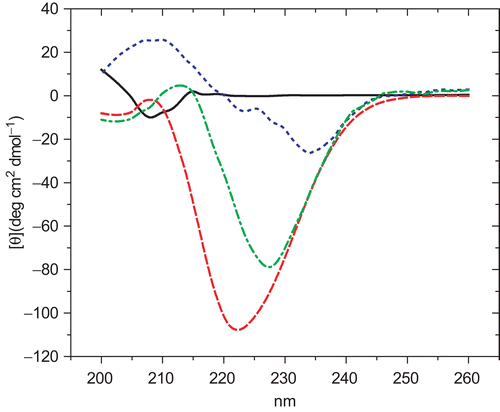
Figure 4. Schematic 1H-13C nuclear magnetic resonance spectra of the methyl groups of the methionine residues in (•) Ca2+-CaM and (○) Ca2+-CaM-1 complex. Chemical shift differences of methionines at the C-terminal (Met109, Met124, Met144, Met145) and N-terminal (Met36, Met51, Met71, Met72) domains of CaM, as well as the Met76 of the flexible linker region, are indicated with solid arrows.
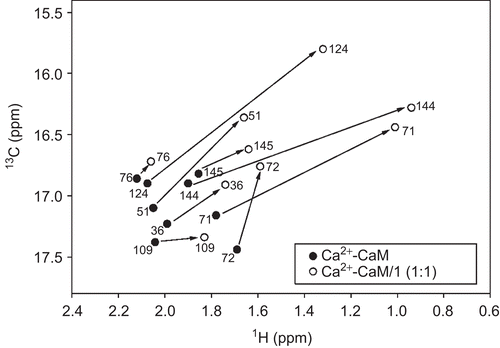
Table 2. NMR and flexible docking results of the Ca2+-CaM−1 complex formation.
Table 3. Binding free energy (EFEB) and inhibition constant (Ki) values of compounds 1–4, obtained from the molecular docking analyses (AutoDock 4.0).
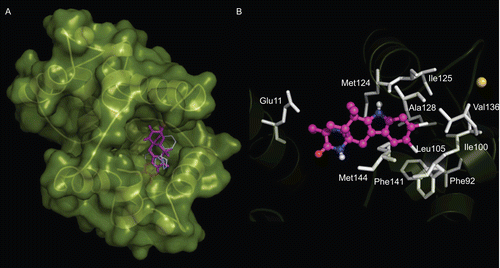
Figure 5. Flexible docking model of the complex Ca2+-CaM−1. CaM is represented in green surface, 1 is depicted in purple sticks, and CPZ is shown in white lines. (A) Lowest energy AutoDock conformation of 1; (B) Residues of CaM (white sticks; Glu11, Phe92, Ile100, Leu105, Met124, Ile125, Ala128, Val136, Phe141, and Met144) involved in the complex formation. Ca2+ ions are showed in yellow balls.