Figures & data
Figure 1. GGT reactions with γ-Glutamyl Substrates. Cleavage of glutathione, a physiologic substrate of GGT, is shown. The γ-glutamyl bond between the γ-carbon of glutamate and the amine of cysteine is cleaved by GGT. The γ-carbon of glutamate forms an acyl covalent bond with the hydroxyl group on the side chain of Thr-381 of human GGT. The acyl bond can either be hydrolyzed releasing glutamate (Glu), or an acceptor nucleophile can attack forming a new γ-glutamyl compound (transpeptidation reaction). The reactions proceeds via a Ping-Pong mechanism.
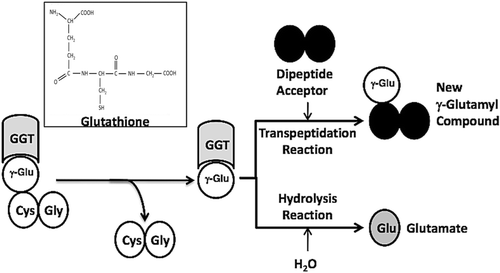
Figure 2. Synthesis scheme for the OU749 analogs. Synthesis and purification of the synthetic intermediates TDA1-10 (A) was required before synthesis of the OU749 analogs Compounds 2–4 and Compounds 6–20 (B). Synthesis of Compound 5 is derived from Compound 11 (C).
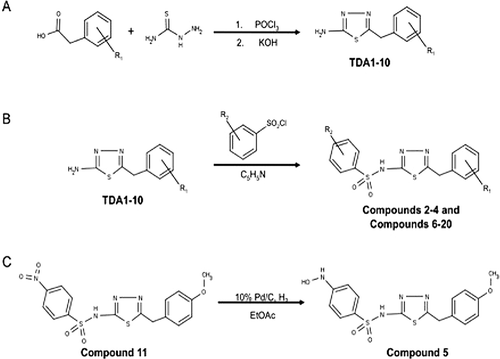
Table 1. Kinetic parameters for reaction with d- and l-GpNA.
Figure 3. Initial velocity pattern with d-GpNA or l-GpNA as the substrate and glygly as an accepter. Initial rates were measured at pH 7.4 and 37 °C. Double recipricol plots of the initial velocity versus the glygly concentration with 0.25 mM (♦),0.5 mM (•),1 mM (▴), or 3 mM (▪) d-GpNA (A) or l-GpNA (B).
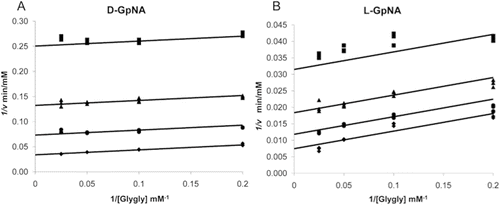
Table 2. Inhibition of GGT by structural analogs of OU749.
Figure 4. Kinetic analysis of GGT inhibition by OU749 and SBS. The structure of OU749 (A). Double-reciprocal plot of the initial velocity of the hydrolysis of d-GpNA in the presence of 0 (♦), 15.2 µM (▾), 31.25 µM (▴), 62.5 µM (▪), 125 µM (•) OU749 (B). The structure of SBS (C). Double reciprocal plot of the initial velocity of the hydrolysis of d-GpNA in the presence of 0 (♦), 6.25 mM (▾), 12.5 mM (▴), 25 mM (▪), 50 mM (•) SBS (D). Double reciprocal plot of the initial velocity of the transpeptidation reaction with varying concentrations of l-GpNA and 40 mM glygly (E) or varying concentrations of glygly with 3 mM l-GpNA (F) in the presence of 0 (♦), 6.25 mM (▾), 12.5 mM (▴), 25 mM (▪), 50 mM (•) SBS. Data shown are average triplicate values ± S.D. For many data points the S.D. is smaller than the symbol.
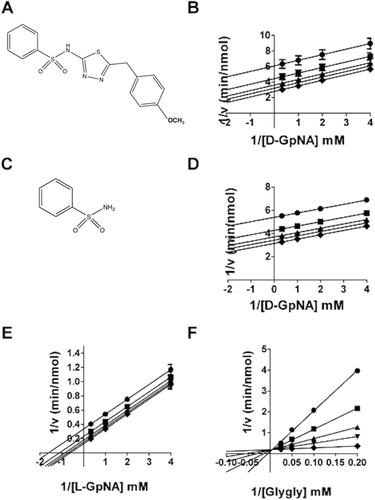
Figure 5. Kinetic analysis of GGT inhibition by Compound 11. Substrate velocity curve of the hydrolysis reaction versus d-GpNA concentration in the presence of 0 (♦), 15.2 µM (▾), 31.25 µM (▴), 62.5 µM (▪), 125 µM (•) Compound 11 (A). Velocity vs Compound 11 concentration (closed symbols) and V/K vs Compound 11 concentration (open symbols) plots illustrate the activation of the hydrolysis reaction (B). Double-reciprocal plots of the initial velocities of the transpeptidation reaction varying l-GpNA with 40 mM glygly (C) or varying glygly with 3 mM l-GpNA (D) in the presence of 0 (♦), 15.2 µM (▾), 31.25 µM (▴), 62.5 µM (▪), 125 µM (•) Compound 11. Data shown are average triplicate values ± S.D.
Table 3. Kinetic parameters for activators of the D-GpNA hydrolysis reaction and Inhibition of the L-GpNA transpeptidation reaction.
Figure 6. Time courses for the hydrolysis of d-GpNA in the presence of acivicin. The release of pNA from 0.287 mM d-GpNA (A) or 2 mM d-GpNA (B) was monitored over time in the presence of acivicin (concentrations of acivicin noted in A). The reactions were conducted at pH 7.4. The increase in the Km of d-GpNA with increasing concentrations of acivicin is shown (insert 6A).
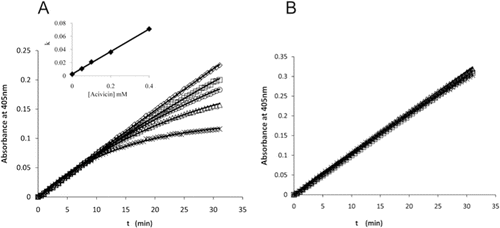
Figure 7. Kinetic analysis of GGT inhibition by Compound 20. Double-reciprocal plot of the initial velocity of the hydrolysis of d-GpNA in the presence of 0 (♦), 15.2 µM (▾), 31.25 µM (▴), 62.5 µM (▪), 125 µM (•) Compound 20 (A). Double reciprocal plot of the initial velocity of the transpeptidation reaction with varying concentrations of glygly with 3 mM l-GpNA (B) in the presence of 0 (○), 15.2 µM (♦), 31.25 µM (▾), 62.5 µM (▴), 125 µM (▪), 250 µM (•) Compound 20. Data shown are average triplicate values ± S.D. For many data points the S.D. is smaller than the symbol.
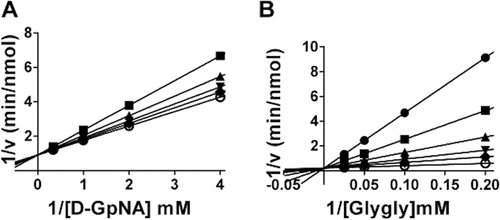
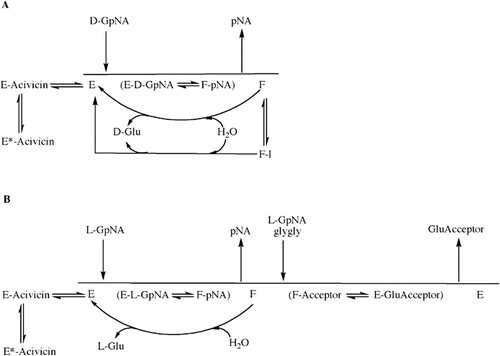
Figure 8. Proposed overall kinetic mechanism of inhibition and activation of GGT. Mechanism of hydrolysis of d-GpNA (A). Enzyme (E) is γ-glutamylated by d-GpNA with release of pNA to give a form of the enzyme (F) that can transfer the γ-glutamyl moiety to water. d-GpNA is not capable of acting as an acceptor. The ester bond can be hydrolyzed with rate constant khyd, and this rate is decreased in the presence of an inhibitor such as OU749 (F-I), or increased in the presence of an activator such as Compound 11 giving rate k’hyd. Acivicin can bind to the free enzyme at the acceptor site, and is slowly trapped (tightly bound) on enzyme. Mechanism of transpeptidation of GpNA (B). Enzyme (E) is γ-glutamylated by GpNA with release of pNA to give a form of the enzyme (F) that can transfer the γ-glutamyl moiety to an acceptor (l-GpNA and/or glygly). The ester bond can be hydrolyzed with rate constant khyd, but competition between water, l-GpNA and/or glygly decreases the rate of hydrolysis. As in panel A, acivicin can bind to the free enzyme at the acceptor site, and is slowly trapped on enzyme.