Figures & data
Figure 1. Michaelis–Menten plot shows that above 30 mM sucrose concentration, the enzyme reaches to zero-order reaction.
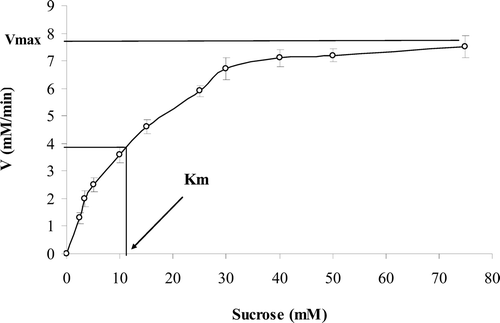
Figure 2. The pattern of Eadie–Hofstee plot showed noncompetitive inhibition after binding of ranitidine to the enzyme. The slope of the lines is the same and determines a constant Km, but intercepts on the ordinate were different, which indicate that the Vmax of the enzyme was not constant in the presence of different concentration of ranitidine (0–3 mM).
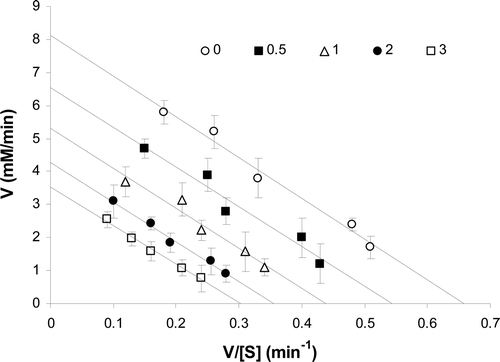
Figure 3. Change in maximum velocity (Vmax) of the enzyme in the presence of different concentrations of ranitidine. Determination of inhibitor concentration yielding 50% inhibition (IC50).
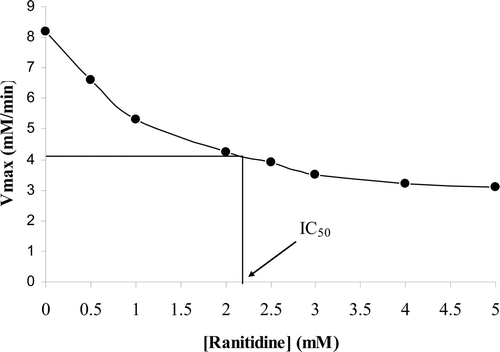
Figure 4. Estimation of Ki by Dixon plot in the presence of different sucrose concentrations (3.3–30 mM).
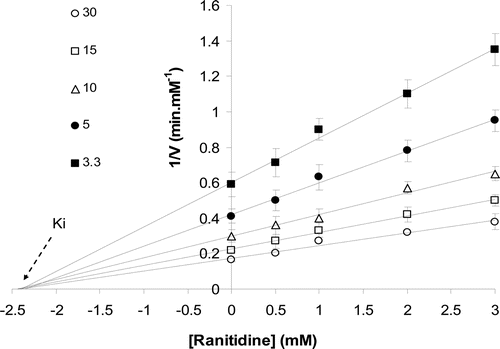
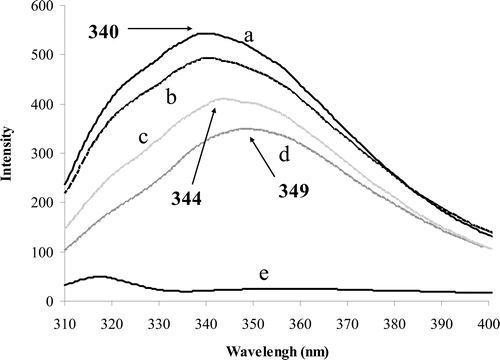
Figure 5. Fluorescence spectra obtained from excitation at 285 nm and emission range of 310–400 nm. The pure enzyme (0.04 g/ml) induces a peak at 340 nm (a). Binding of substrate (30 mM) to the enzyme reduced the peak emission intensity (b). Binding of ranitidine (6.5 μM) to free enzyme also decreased the emission intensity and induced a red shift to 344 nm (c). Binding of ranitidine to enzyme–substrate complex also induced red shift (d). Pure ranitidine (e).