Figures & data
Figure 1. Chemical structures of D/L-Ile-Phe-Lys, YO-2, and PKSI-527. KM and IC50 values for plasmin are in parenthesesCitation3,Citation4,Citation20.
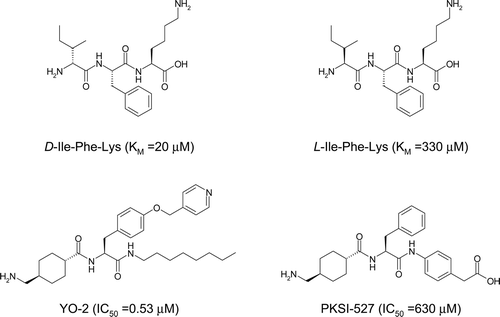
Figure 2. Molecular dynamic simulation of plasmin over 11 ns. The root-mean square deviation (RMSD) of simulated structures from the initial structure is plotted versus time. The RMSD calculation is based on the protein main-chain atoms.
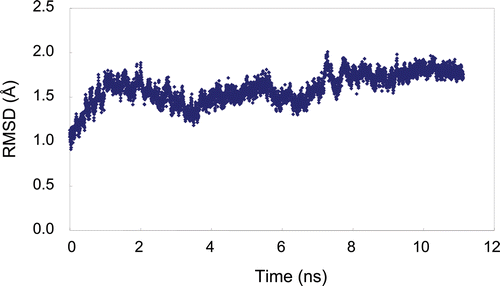
Figure 3. Predicted complex of trypsin with PKSI-527. The structure of trypsin is displayed as a cartoon model (grey) and the inhibitor PKSI-527 is shown as a stick model (yellow). The side-chains of His57, Asp102, Asp189, and Ser195 in the catalytic site of trypsin are shown as stick models (brown).
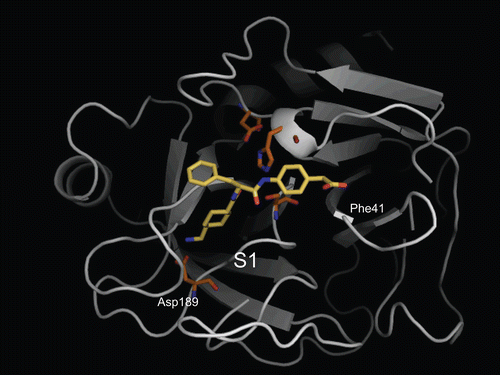
Figure 4. Predicted complex of plasmin with D/L-Ile-Phe-Lys substrates. The structure of plasmin is displayed as a surface model (grey) and the ligands D- and L-Ile-Phe-Lys are shown as stick models (magenta for D-Ile-Phe-Lys and blue for L-Ile-Phe-Lys). The side-chains of residues in the binding and catalytic sites are shown as stick models (white). The underlined label is for the substrate. (a) Close-up view of S1/P1 interaction. (b) The overall binding mode of the substrates.
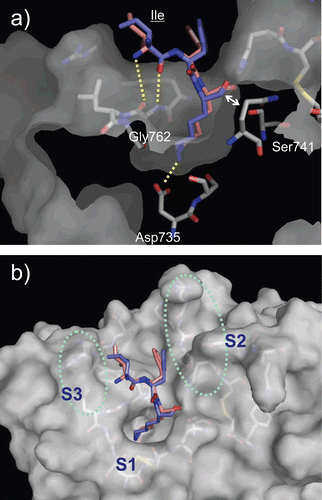
Figure 5. Predicted complex of plasmin with YO-2. The structure of plasmin is displayed as a surface model (grey) and the inhibitor YO-2 is shown as a stick model (green). The side-chains of residues in the binding and catalytic sites are shown as stick models (white). (a) Identical view as that of . (b) Side-view.
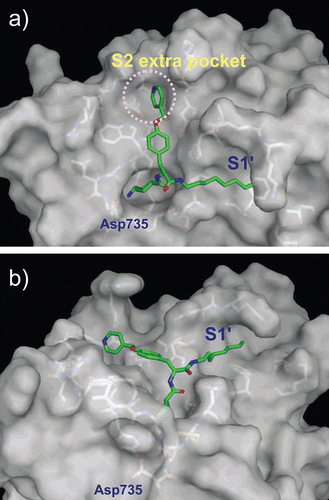
Table 1. SAR of Tra-Tyr(O-Pic)-CONH-X.
Figure 6. Predicted complex of plasmin with PKSI-527. The structure of plasmin is displayed as a surface model (grey) and the inhibitor PKSI-527 is shown as a stick model (yellow). The side-chains of residues in the binding and catalytic sites are shown as stick models (white). (a) Identical view as that of . (b) Comparison of the binding modes of YO-2 (green) and PKSI-527. (c) The structure of PKSI-527 overlapped with YO-2.
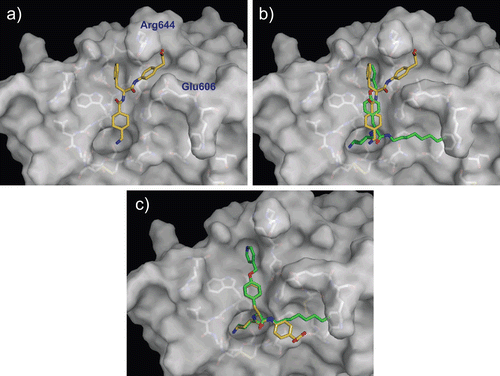
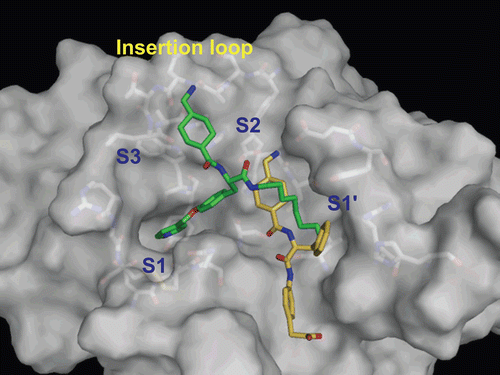
Figure 7. Predicted complex of urokinase with YO-2 and PKSI-527. The structure of urokinase is displayed as a surface model (grey) and the inhibitors YO-2 and PKSI-527 are shown as stick models (green for YO-2 and yellow for PKSI-527). The side-chains of residues in the binding and catalytic sites are shown as stick models (white).