Figures & data
Table 1. Crystal data and diffraction data collection and refinement statistics.
Figure 1. (A) Overall three dimensional structure and secondary structure elements of Sapp1p complexed to RTV. The protein in the ribbon representation is colored by secondary structure, the RTV molecule is represented by sticks. the transparent protein solvent accessible surface area is also shown. (B) The 2Fo-Fc electron density map for RTV is contoured at 0.8σ. The carbon atoms of RTV are shown in green, oxygen, nitrogen and sulfur atoms are colored red, blue and golden, respectively. Two catalytic aspartates are depicted with carbon atoms colored yellow. (C) The detail of the RTV interaction with residues in the Sapp1p active site. Hydrogen bonds are shown as dashed lines; the number represents distance between hydrogen bond donor and acceptor in Å.
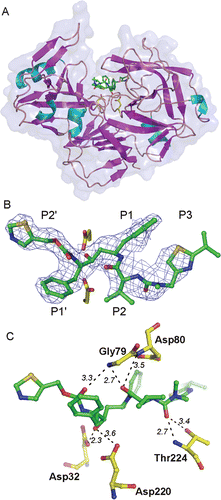
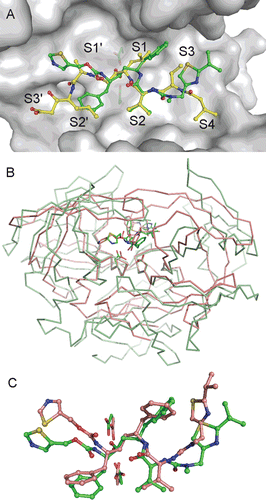
Figure 2. (A) Comparison of RTV and PepA binding modes in the Sapp1p active site. Carbon atoms of RTV and PepA are shown in green and yellow, respectively. Oxygen, nitrogen and sulfur atoms are colored red, blue and golden, respectively. Protein is shown by a solvent accessible surface area with underlying catalytic aspartates shown as sticks. Substrate binding sites S4-S3′ are labeled. Flap region (residues 71–89) is omitted from the figure for clarity. (B) Superposition of Sapp1p and HIV-1 PR complexes with RTV. Structure of HIV-1 PR in complex with RTV (PDB code 1HXWCitation16) is colored pink; the structure of Sapp1p in complex with RTV is colored green. Superposition was done by matching the catalytic triads (catalytic aspartates are highlighted as sticks at the bottom of the active sites). (C) Comparison of RTV binding modes in the active sites of Sapp1p (carbon atoms colored green) and HIV-1 PR (carbon atoms colored pink).