Figures & data
Figure 1. (A) Schematic map of the construct on the recombinant plasmid pMSH-3-(OXA-162). The coding regions are shown as arrow-head boxes indicating the direction of open reading frame. The insertion sequence IS1R was shown as gray-shaded box. (B) The left insertion sequence IS1999 is disrupted by IS1R between the dedicated promoter signal sequences while leaving the -10 signal intact.
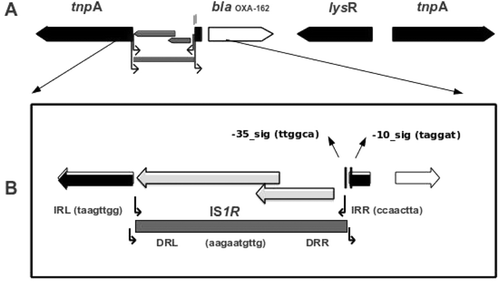
Table 1. Kinetic parameters for OXA-48 and OXA-162.
Table 2. Minimum inhibitory concentration (MIC) of OXA-162 producing K. pneumoniae strain KIRK1, its transformant, OXA-162 producing clone and OXA-48 producing clone.
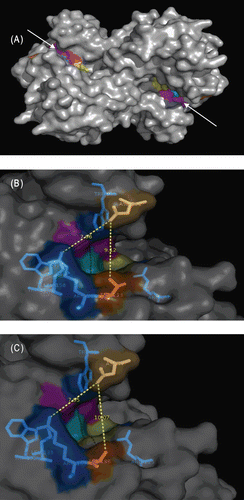
Figure 2. Examination of the active site regions of OXA-48 and modeled OXA-162. (A) OXA-48 homodimer (active site entry regions were pointed out with arrow heads and active site residues were colored). (B) Depiction of OXA-48 entry site. Dark orange colored amino acid (Thr213) is where the mutation occurred. Asp101 was located across the mutated amino acid. The measured distance between Thr213 and Asp101 was 9.12 Å. This was predicted to be the putative antibiotics entry site. (C) Depiction of the modeled OXA-162 entry site. Dark orange colored amino acid is the mutated amino acid (Ala188). Asp77 was located across the mutated amino acid. The measured distance between Ala188 and Asp77 was 10.07 Å which indicated the presence of 1 Å widening of the predicted entry site. Note that the corresponding amino acids for Ala188 and Asp77 in OXA-48 were Thr213 and Asp101, respectively.