Figures & data
Table 1. The strain codes, clades, sequences, TCID50 values (median tissue culture infective dose) and percentages of neutralization in soluble CD4 (sCD4) (10 µg/mL) of the CD4-binding sites for the 60 gp120 strain panel considered.
Figure 1. Homology modelling of free gp120 corresponding to strain code AY71341 (sequence number 11 out of the 60 strains considered). The 41 amino-acids of the CD4-binding site are bolded and underlined. The alignment follows Chen et al.Citation2.
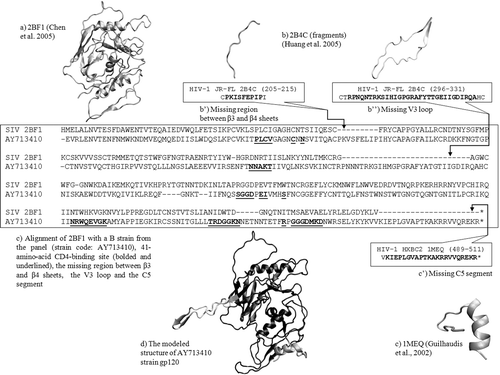
Table 2. Significant structural, energetical and sterical descriptors of the CD4-binding sites for the 60 gp120 strains and their statistics.
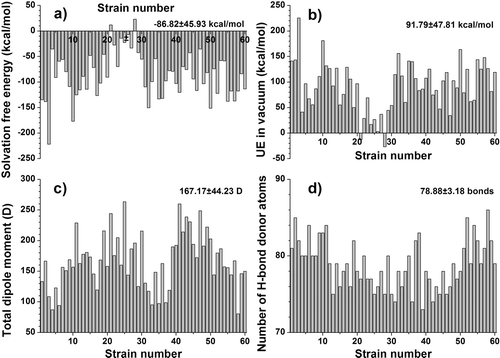
Figure 2. Variation of significant structural descriptors of the CD4-binding sites for the 60 gp120 strains. (a) SAS of positive atoms; (b) SAS of and negative atoms; (c) SAS of hydrophobic atoms; (d) SAS of polar atoms. The mean values and the standard deviations are noted in the higher right corner. The Y-axes are adjusted for a clearer representation of the descriptor variation.
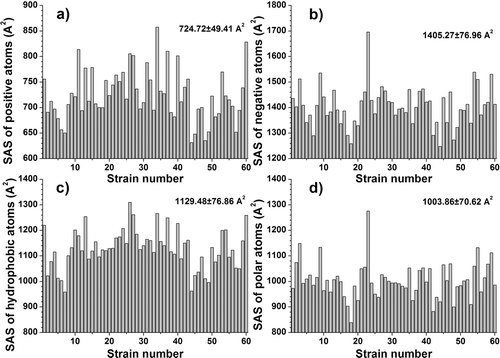
Figure 3. Variation of significant energetical and sterical descriptors of the CD4-binding sites for the 60 gp120 strains. (a) free solvation energy; (b) electrostatic surface potential energy (UE) in vacuum; (c) total dipole moment; (d) the number of hydrogen-bond donors. The mean values and the standard deviations are noted in the higher right corner. The Y-axes are adjusted for a clearer representation of the descriptor variation.