Figures & data
Figure 2. 1H NMR spectrum of the complex Ag(tpp)3(asp) in DMSO-d6: (top) aliphatic region and (bottom) aromatic region.
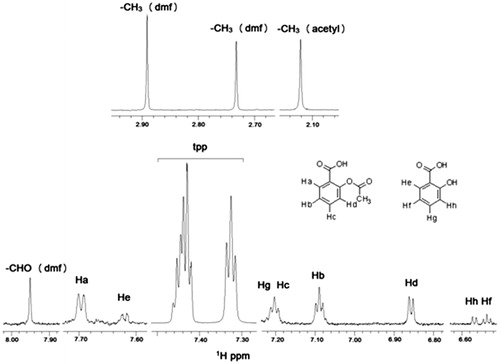
Figure 3. 1H NMR spectrum of soybean LOX-1 with complex Ag(tpp)3(asp) in Tris/D2O: (top) without sonication and (bottom) after sonication.
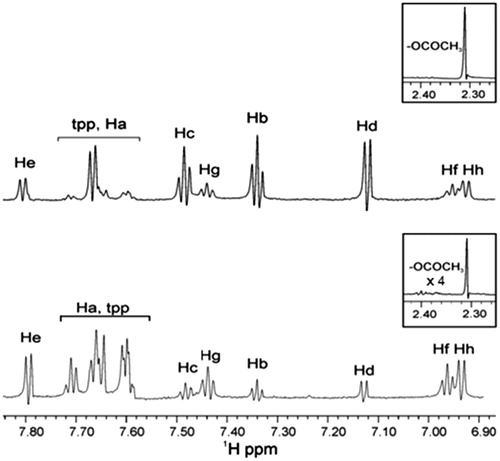
Figure 4. STD experiment of 1H NMR of soybean LOX-1 with complex Ag(tpp)3(asp) in Tris/D2O (without sonication): the reference NMR spectrum of aliphatic and aromatic region (top), and the STD NMR spectrum of aromatic region (bottom).
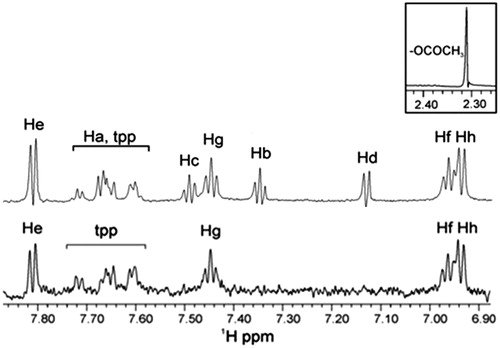
Figure 5. STD 1H NMR of soybean LOX-1 with complex Ag(tpp)3(asp) in Tris/D2O (after sonication): the reference NMR spectrum of aliphatic and aromatic regions (top), and on-resonance STD NMR spectrum of aromatic region (bottom).
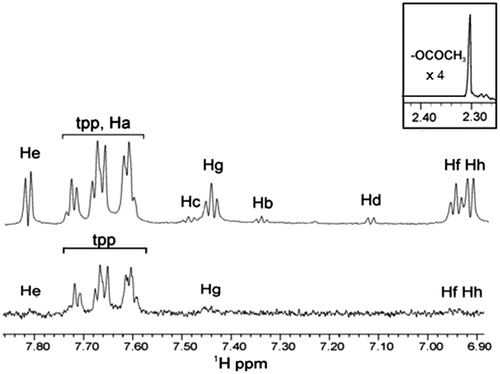
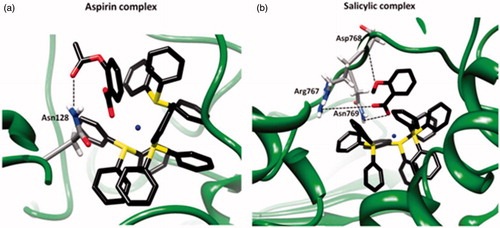
Figure 6. Ligand map (dotted lines: hydrogen bonds) of the complex of (a) aspirin and (b) salicylic acid in the cavity of LOX-1 (1F8N).
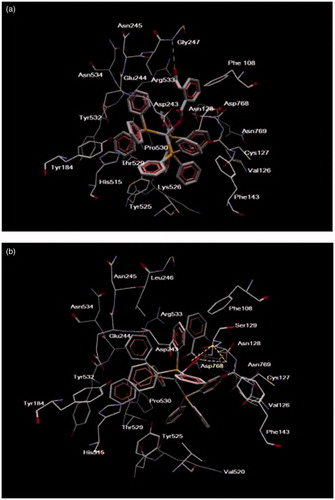
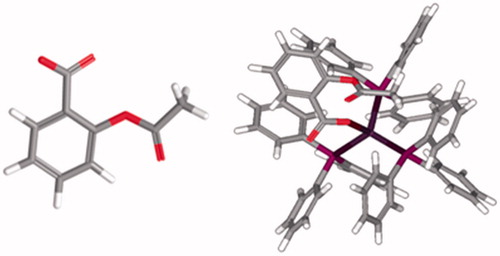
Figure 7. The Ag complex inside binding cavity of LOX-1. The atom of Fe(II) in the active site of cavity 1 is also displayed.
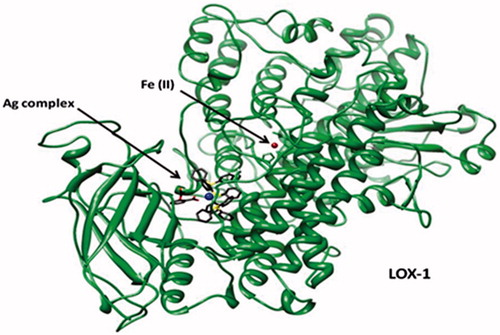
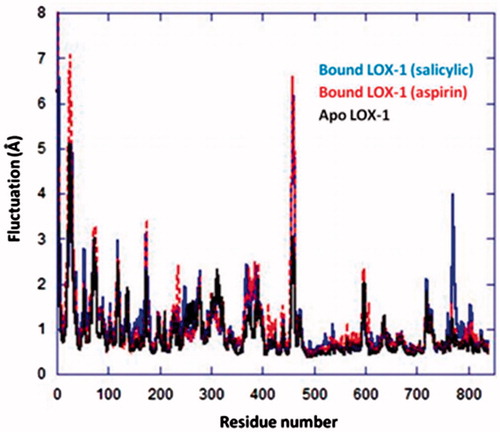
Figure 8. RMSD of LOX-1 bound to (a) Ag complex (salicylic) and (b) Ag complex (aspirin). RMSD values for the apo form of LOX-1 and for the two Ag complexes in the bound-protein systems are also shown. Calculations were initiated from each structure obtained after docking and overlapped on the same structure. For LOX-1 and Ag complexes, superposition involved Cα atoms and all heavy atoms, respectively.
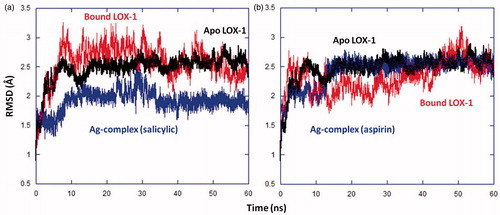
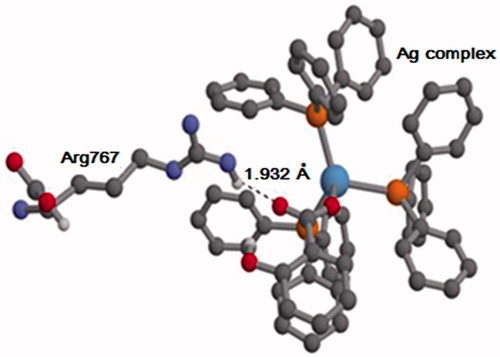
Figure 10. Hydrogen bonds between Ag complex and residues in binding cavity of LOX-1 for (a) the aspirin complex and (b) the salicylic complex. Interactions are represented by dotted lines, and involve oxygen atoms (red) in the aspirin/salicylic moiety of Ag complex. Ag(I) and P atoms are shown in blue and yellow, respectively; for simplicity, the hydrogen atoms of Ag complex are not shown.