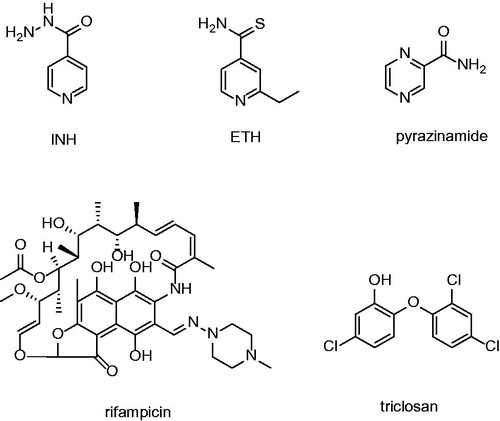
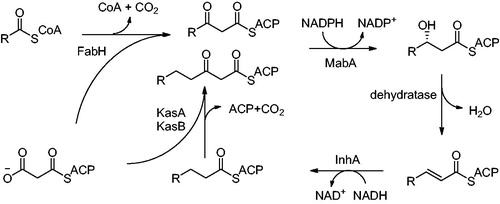
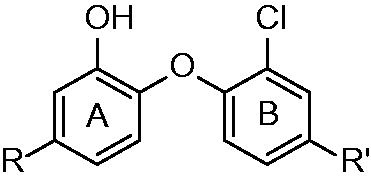
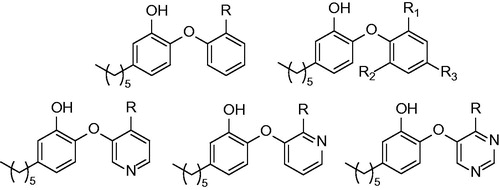
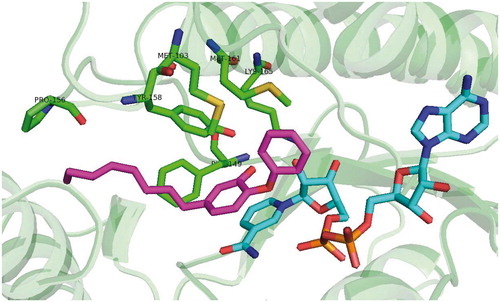
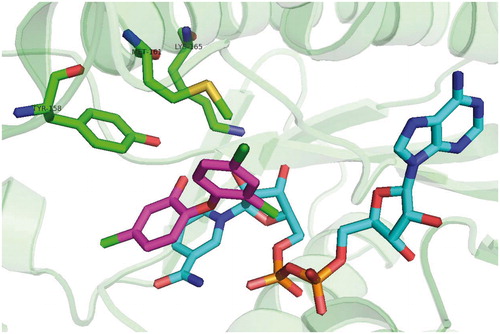
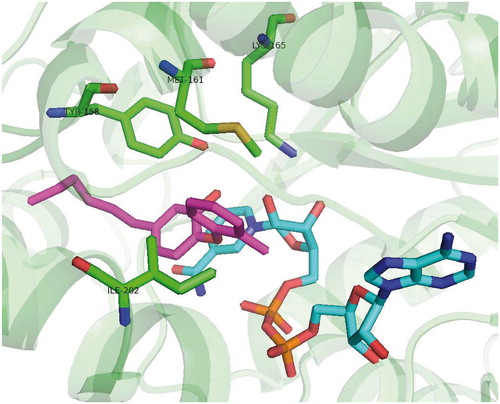
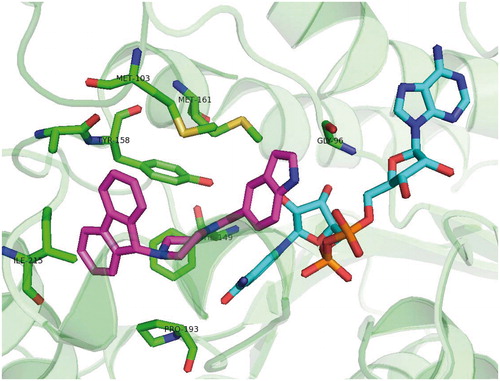
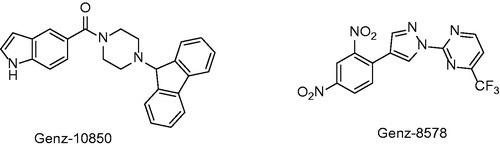
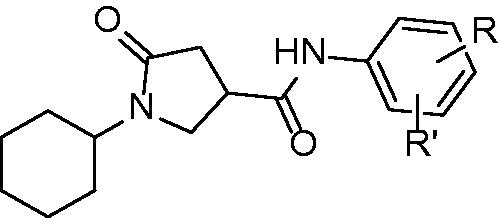
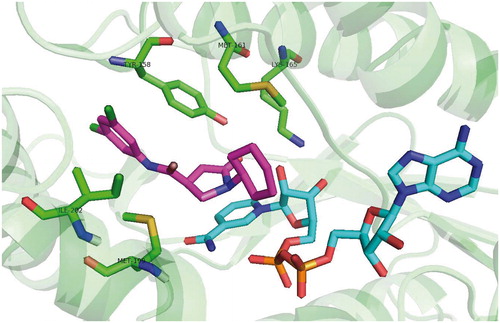
Figures & data
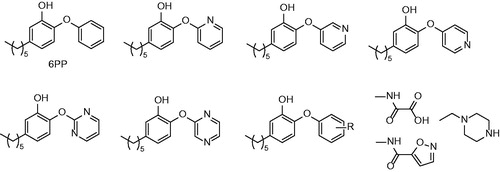
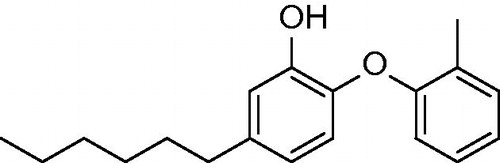
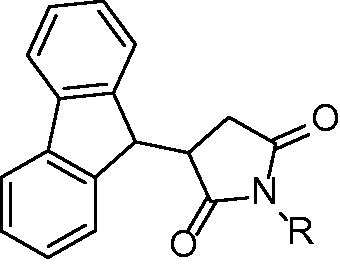
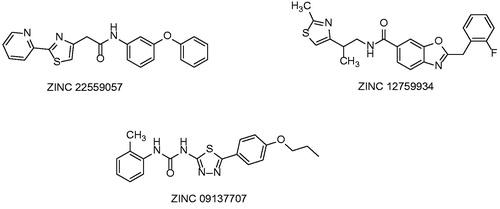
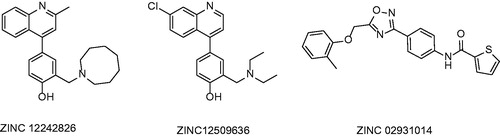
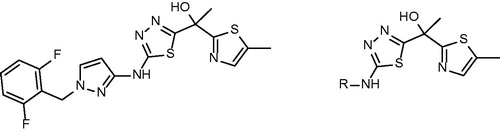
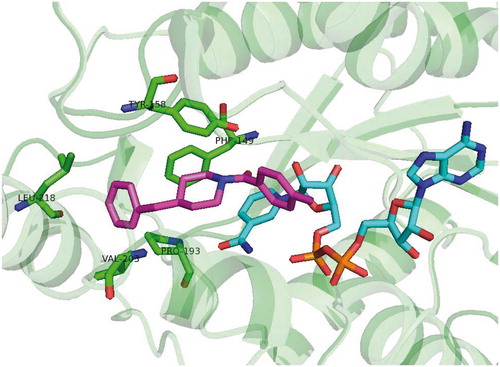
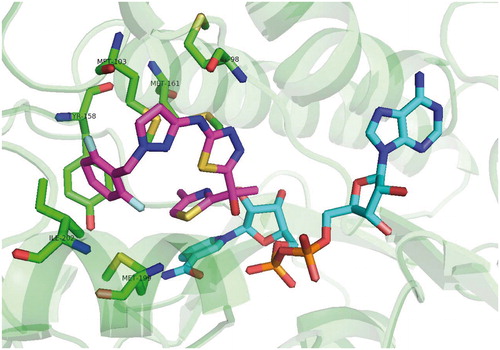
Table 1. Structures and in vitro results of notable InhA inhibitors.