Figures & data
Table 1. Active ERβ agonists used in the enriched dataset.
Figure 2. Results of the cross-docking study. The average RMSD (A) and the number of poses with a RMSD less than 2 Å (B) is reported.
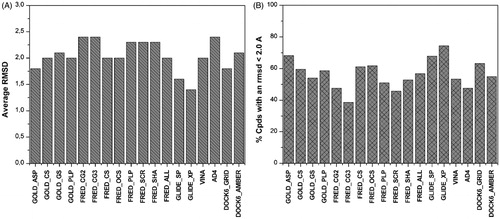
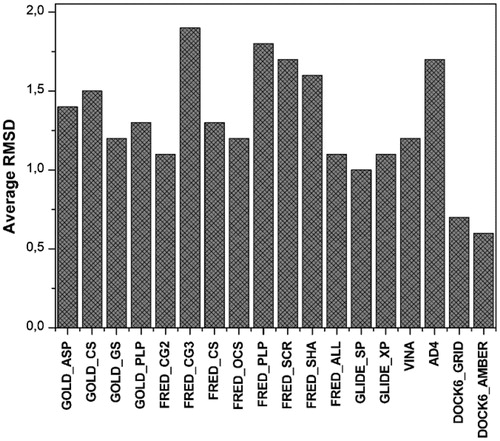
Figure 3. AUC (black) and EF10% (white) values of the first enriched dataset using each scoring function.
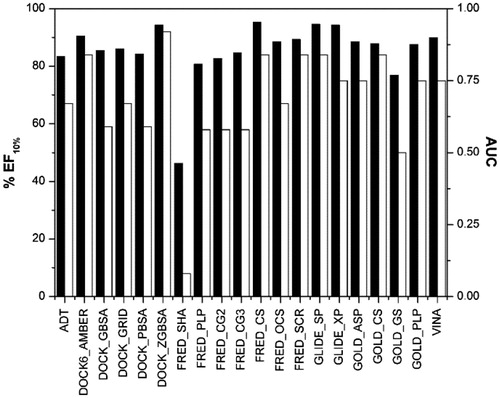
Figure 4. AUC (black) and EF10% (white) values of the second enriched dataset using each scoring function.
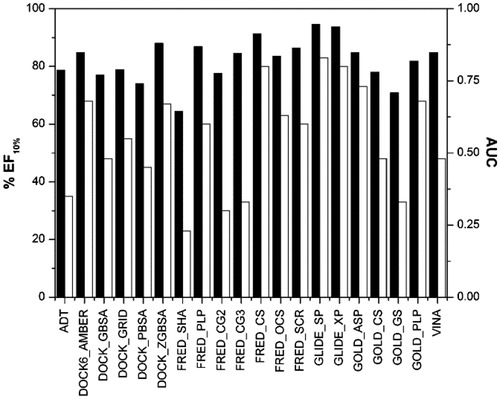
Figure 5. Virtual screening workflow. *The compounds that after the docking calculation showed the two H-bonds with H475 and the E305-R346 network system were taken into account. **The compounds that maintained the H-bonds with H475 and the E305-R346 network system for at least the 75% of MD simulation were considered.
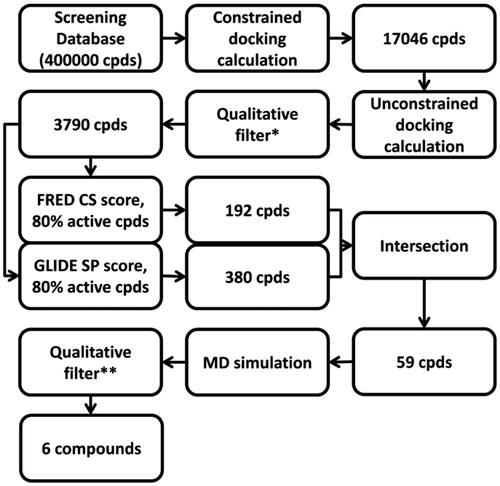
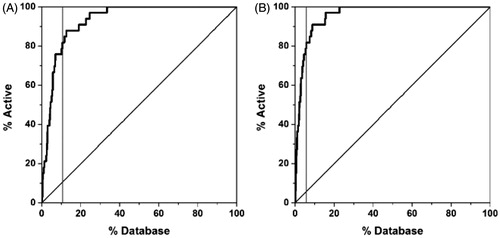
Figure 6. Rescoring results for the enriched database using GLIDE (A) and FRED (B). The grey vertical line corresponds to the threshold used for filtering the compounds.