Figures & data
Figure 1. The reactions catalyzed by PHM and PAL (A), the cinnamate-derived vinyl radical (B) and the 4-phenyl-3-buteneoate-derived allylic radical (C). Bifunctional PAM is compromised of separate monofunctional enzymes, PHM and PAL. In vivo, the substrate for PHM and PAL is a C-terminal glycine extended peptide (R = a peptide) to generate an α-amidated peptide.
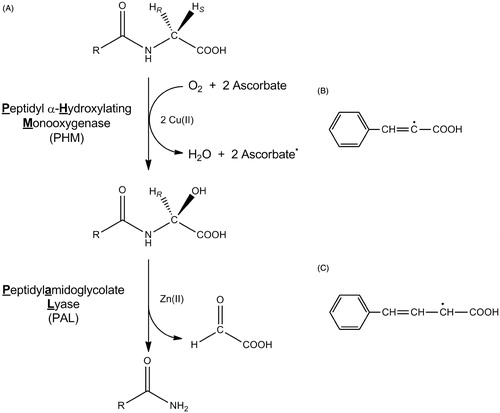
Figure 2. Inhibition of PHM by cinnamic acid. Assays were performed as described in “Methods” section for the initial rate determination. The points are initial rates and the lines were computer fit using SigmaPlot 9.0. Initial rates of O2 consumption were measured for ≤3 min, during which the inactivation mediated by cinnamate makes a neglible contribution to the observed inhibition.
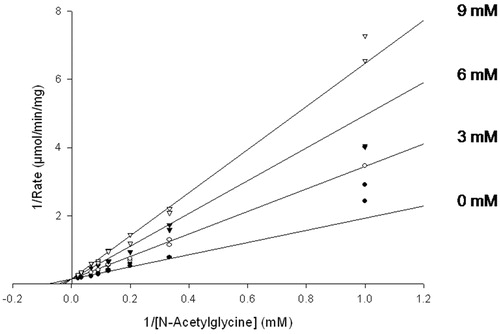
Table 1. KI,obs values for the inhibition of PHM by cinnamate and the cinnamate analogsa.
Figure 3. Inactivation of PHM by cinnamic acid. (A) PHM was incubated at 37 °C in the presence of the indicated concentration of cinnamate as described in the “Methods” section. At the indicated times, an aliquot was removed and assayed for residual O2 consumption activity. Lines are linear regression fits to the equation: log[(vt/v0)] = (−kobs/2.303)t + C, where v0 is the average initial rate of O2 consumption in the absence of cinnamate, vt is the initial O2 consumption rate at time = t, and C is a constant which should be within experimental error of 2.0. The error bars represent the standard deviation of the duplicate measurements. (B) A reciprocal replot of the inactivation data from panel A.
![Figure 3. Inactivation of PHM by cinnamic acid. (A) PHM was incubated at 37 °C in the presence of the indicated concentration of cinnamate as described in the “Methods” section. At the indicated times, an aliquot was removed and assayed for residual O2 consumption activity. Lines are linear regression fits to the equation: log[(vt/v0)] = (−kobs/2.303)t + C, where v0 is the average initial rate of O2 consumption in the absence of cinnamate, vt is the initial O2 consumption rate at time = t, and C is a constant which should be within experimental error of 2.0. The error bars represent the standard deviation of the duplicate measurements. (B) A reciprocal replot of the inactivation data from panel A.](/cms/asset/bdc0b08a-df68-4d82-81b3-e4bbd1b729bb/ienz_a_1046064_f0003_c.jpg)
Table 2. Values for kinact,obs and (kinact/KI)obs for cinnamate and selected cinnamate analogs.
Figure 4. MALDI-TOF overlay of trypsin-digested active and cinnamate-inactivated PHM. The catalytically active control is shown in blue, while the inactivated PAM is shown in red. Inactivation was achieved through incubation with cinnamate as described in the “Methods” section. This figure is best viewed in color, which is only available online.
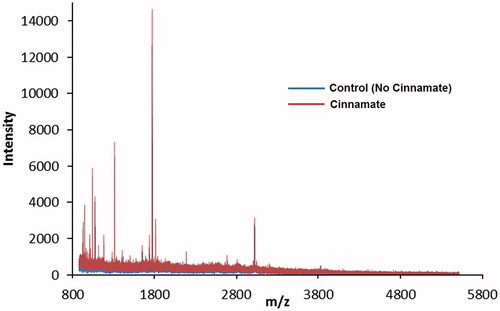
Figure 5. Binding of cinnamate into the PHM active site (A) and the relative orientation of cinnamate to bound substrate (N-α-acetyl-3,5-diiodotyrosylglycine, IYT) in PHM (B). The orientation of both the IYT (blue) and cinnamate (red) ligands is superimposed to compare recognized electrostatic (Arg-240) and hydrophobic (orange mesh) interactions near the CuM domain associated with productive ligand bindingCitation53,Citation62. PHM contains two bound copper atoms, CuM and CuHCitation53.
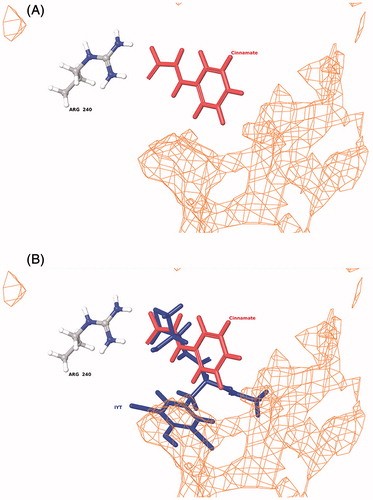
Table 3. Relative bond dissociation energies calculated for selected PHM inhibitors or inactivators.
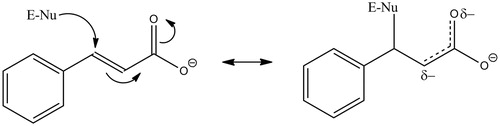
Figure 6. Proposed mechanism for the cinnamate-mediated inactivation of PHM. E-Nu represents an active site nucleophile, possibilities include Lys-134, Glu-313, Tyr-318 Met-109 or Met-314.