Figures & data
Table 1. Bacterial strains used for mouse inoculation with their respective growth media and primers used for real-time PCR enumeration
Figure 1. Experimental set up. Five groups of mice (males in groups A–D; females in group E) were individually caged and distributed into 5 isolators. Different bacterial suspensions (mixes or pure cultures) were inoculated at several time points between days 0 and 14 (see also ). On day 14, mice from groups D and E were removed from their individual cages and mixed with the other mice from the same group. On day 56, 3 GF mice were introduced into the isolators containing groups D and E. All animals received standard chow diet. p.i., post-inoculation.

Table 2. Protocol for the second mouse inoculation experiment
Figure 2. Colonization pattern in mice from groups A and D. Individual bacterial strains were quantified by real-time PCR in feces collected between days 3 and 70 post-inoculation and values are expressed as log10(cells) g−1 of feces. Dots represent bacterial counts in individual mice. Mean values are shown as lines between dots. The detection limit is approximately 106 cells g−1.
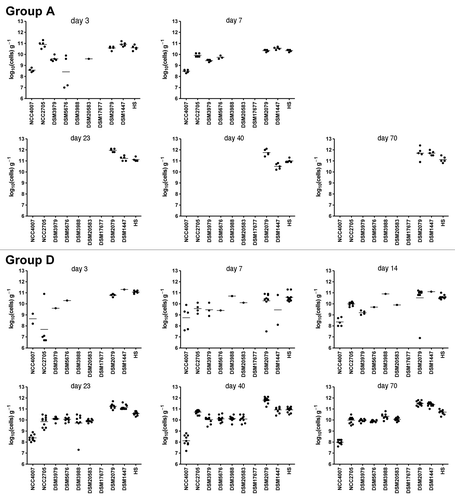
Figure 3. Transfer of the simplified microbiota from colonized mice to GF mice. Three GF male mice were introduced in the isolator containing mice from group D on day 56 post-inoculation. Individual bacterial strains were quantified by real-time PCR in feces collected at days 2, 7 and 14 after transfer of the GF mice and values are expressed as log10 (cells) g−1 of feces. Numbers in parenthesis indicate day post-inoculation of mice in group D. Dots represent values from individual mice. Mean values are shown as lines between dots. The detection limit is approximately 106 cells g−1.
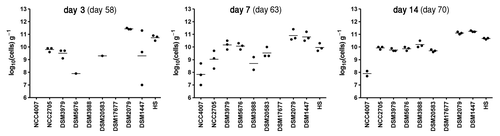
Table 3. Summary of variations of plasma (P) and urinary (U) metabolite signals obtained by pairwise comparisons between animals harboring 3 (pooled data from groups A, B and C), 9 (group D; males) and 8 (group E; females) bacterial strains
Figure 4. O-PLS-DA cross-validated scores plot derived from 1H NMR spectra of urine (A) and plasma (B) and (C) [NOESY (B) and CPMG (C) acquisition] from mice harboring three bacteria (groups A, B and C; males), nine bacteria (group D; males) and eight bacteria (group E, females).
![Figure 4. O-PLS-DA cross-validated scores plot derived from 1H NMR spectra of urine (A) and plasma (B) and (C) [NOESY (B) and CPMG (C) acquisition] from mice harboring three bacteria (groups A, B and C; males), nine bacteria (group D; males) and eight bacteria (group E, females).](/cms/asset/ae5aef68-a352-4401-9244-919d5166ac7c/kgmi_a_10918754_f0004.gif)