Abstract
Leishmania is a neglected protozoan parasite which creates some problems for public health with different clinical infections in different countries around. Due to the lack of an effective drug without side effects and the emergence drug resistance, there is an urgent need to introduce the novel drug targets and new drugs and vaccines to control leishmaniasis during recent years, metabolomics and other “Omics” platforms has become an important approach to comprehensive knowledge of the Leishmania parasites biology. The study of metabolite profiles can open the insights for discovering novel therapeutic targets in this infection in both of the parasites and human host. In addition, specifying the metabolomics profile changes among promastigotes, amastigotes and during metacyclogenesis can pay the way for achieving parasite survival parameters and the host-parasite interaction. The previous studies in this field have been extracted from the databases, literature and their detailed major concepts. The present review highlights the role of metabolomics approach in the field of Leishmania research. Also, several important metabolite signatures introduced in various aspect of leishmania parasite such as drug resistance and parasite biology which would be useful in the field of biomarker and drug discovery process. Finally, metabolomics plays a potential role in introducing metabolic pathways related to Leishmania parasite and its treatment design.
1 Introduction
Personal health is considered as one of the principles of health care in the future. The system biology plays a significant role in understanding the interactions of biological system at different levels such as genomes, proteins, and metabolites. Recently, “Omics” approaches including genomics, proteomics, and metabolomics analysis and also bioinformatics and computational prediction are from the modern technologies which have gained much attention in recent years to find potential markers for different diseases using various biological samples such as tissues, biological fluids and cell cultures.Citation1,Citation2 As one of the components of the system biology, metabolomics is also rapidly developing, which has attracted a lot of attention in various biomedical studies. In addition, the metabolomics research has a great potential for coming into the medical arena.Citation3–Citation5 Metabolomics analyzes the whole metabolites which are present within an organism in a specific condition. Through detecting and quantifying all metabolites in a specific sample, metabolomics provides a “snapshot” during the metabolic changes related to the disease.Citation6 In fact, metabolomics studies reflect changes in the upstream stages of system biology research (genome and proteome). Due to more dynamical status of metabolomics rather than both genomics and proteomics analysis, this approach can detect metabolic changes associated with different physiological states within a shorter time frame.Citation3,Citation7 A large body of research emphasized the importance of metabolites as invaluable biomarkers for diagnosing and detecting disease. However, metabolomics can detect and introduce biomarkers in a broad range of samples such as whole blood, serum, plasma, urine, saliva, and the like in various disease conditionsCitation8
2 Leishmania and leishmaniasis
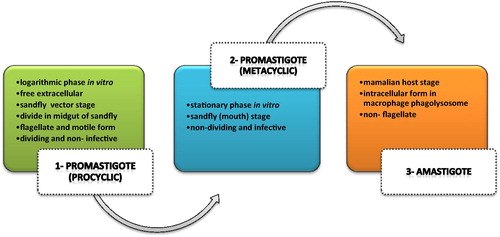
Protozoan Leishmania causes a wide spectrum of clinical disease among humans, which is globally regarded as a major public health problem in the Middle-East.Citation9 Leishmaniasis is a vector-borne disease which is transmitted by sandflies and caused by obligate intracellular protozoan of the genus Leishmania. Human infection appears by about 20 species of parasite which are transformed to human through biting by sand-fly vector.Citation10 Different species are morphologically the same although they are different by isoenzyme analysis. These species include the L. donovani complex with two main species such as L. donovani, L. infantum, the L. mexicana, L. tropica, L. major and L. aethiopica and the subgenus L. viannia with four main species. It is estimated that over 350 million people are at risk for leishmaniasis, among whom 12 million were infected and 1.5–2 million new cases are globally reported each year.Citation9,Citation11 There are several different forms of leishmaniasis in the human based on species-dependent disease, ranging from self-healing skin ulcers in cutaneous leishmaniasis to the most severe pathologies associated with visceral leishmaniasis by L. donovani, L. infantum and L. chagasi, which is fatal if it is not treated appropriately.Citation12 Cutaneous leishmaniasis is the most common form of leishmaniasis and L. major and L. tropica are the main cause for this form of leishmaniasis making skin lesions.Citation13 The Leishmania parasite has a dimorphic life cycle including an infective promastigote form inside the gut of the sand-fly, which are transmitted with saliva into a human host during blood feeding. Within the mammalian host, promastigotes are rapidly engulfed by phagocytic cells at the infection site and undergo cyto-differentiation to the amastigote stage within the phagolysosome of the host macrophage with acidic environment . The differentiation of the promastigote form into the amastigotes is determined by several morphological and biochemical changes which are essential for surviving and propagating parasite during accommodation in phagolysosome vacuoles. Interestingly, promastigote forms can be easily cultured in vitro and the amastigotes of some Leishmania species can be proliferated in cell free mediumCitation14,Citation15 Thus, most studies were conducted on the promastigote form and axenic amastigote and in the meantime, metabolomics studies also are no exception.
Fig. 1 Different forms of Leishmania parasites during their infectious cycle. Procyclic promastigotes exist in the gut of sand-fly vector and divide quickly. Then this form develops into the metacyclic promastigotes forms. Transformation of procyclic to metacyclic is named metacyclogenesis process. In mammalian hosts metacyclic transforms in to intracellular amastigote forms in macrophage cells.
3 Metabolomics
The metabolomics is a part of “Omics” science in system biology which has been developing for understanding the alteration of molecular level in disease conditions. Metabolomics, along with other “Omics” platforms such as genomics, transcriptomics and proteomics, can complete the systemic view of biology in different research areas.Citation16 Metabolomics is defined as an informative analytical tool for metabolites measurement in both quantitative and qualitative conditions.Citation17 Regarding “Omics”, metabolomics is located at the downstream of others which make the system-level rich by metabolites, their concentration assessment, and flux through metabolic pathways. The development of metabolomics and conducting a large body of research in this area are regarded as some evidence of the progression and applicable presence of metabolomics in many areas.Citation18 Identification of the changes in metabolites level can help to diagnose disease in early stages. Metabolites are small endogenous molecules and the end products of biochemical pathways with low molecular weight between 1500 Da and less such as lipids, peptides, organic acids, nucleic acids, amino acids, vitamins and carbohydrates which metabolomics allow for profiling the metabolites globally in various biological samples such as blood, tissue, urine and the like.Citation19 The metabolites are invaluable small molecules playing an important role in biological systems and representing useful data to understand disease phenotypes.Citation20 In addition, profiling metabolite markers and their amounts is regarded as an important method for diagnosing the disease in early stages. Comparative metabolomics by metabolite profiling from different conditions (e.g. disease vs. control) resulted in detecting metabolic changes and supporting the inference of different aspects of the disease such as molecular mechanisms, biochemical pathways, disease prediction or progression, and even screening markers of early diagnosis.Citation21 Metabolites variation levels are applied as the prediction of disease before clinical symptoms of disease. Regarding the metabolomics field, targeted and non-targeted approaches are available as two main categories. Based on targeted approach, specific and known groups of metabolite are associated with certain metabolic pathways while non-targeted approach focuses more on all possible metabolite analyses in any sample in certain condition. Thus, the second approach is preferred.Citation22 Biomarker discovery for various diseases and integrative system biology are considered as the most common areas for metabolomics analysis. Biomarker discovery by using metabolomics tool is a promising approach for disease diagnosis in the early stage before appearing the related symptoms. A comprehensive profile of metabolites in biological specimen is defined as metabolomics.Citation21 In addition, a large number of techniques are used for profiling metabolites, among which we can refer to one dimensional proton nuclear magnetic resonance (1H NMR) and mass spectrometry (MS) as the most commonly used techniques. The development of metabolomics-based studies usually relies on the analytical techniques such as chromatography separation methods, integrated with MS.Citation23 Based on MS-based metabolomics experiments, the quantitative analysis of metabolites is performed with high sensitivity while using sample complexity reduction prior to detection can simplify the identification of metabolites based on LC-MS approach. The type of biological sample is very important in choosing the technique in metabolomics. Recently, the detection of metabolites and their metabolic pathways is made possible by advanced techniques-based MS such as GC-MS, HPLC-MS, UPLC-MS and CE-MS. Each of these applied analytical platforms for detecting metabolites has its own advantages and disadvantages. Thus, these platforms cannot individually detect all metabolites in biological samples. There are metabolome analyses of many types of organisms conducted for obtaining information about metabolic pathways. Therefore, an appropriate metabolomics analysis is required to obtain full understanding of Leishmania metabolic pathways in different aspects of this parasite. The quality of sample preparation is very important for achieving high quality results. This step should be highlighted in different resources of biological samples in Leishmania and an individual protocol is necessary for assessing the metabolome profile of cultured parasite (∼108 cells) by NMR or MS-based methods .
Fig. 2 The metabolomics-based study of Leishmania parasite process for NMR and LC-MS analysis. Sample preparation step of cells for NMR and MS-base analysis is different from each other which applying appropriate protocol could be obtain better results. The study of the metabolome of Leishmania-infected cells using metabolomics, can lead to the identification of metabolite targets for chemotherapeutic treatment and introducing of novel molecular markers that can be used for the diagnosis of different diseases.

3.1 Analytical approach in metabolomics
3.1.1 NMR (Nuclear Magnetic Resonance)
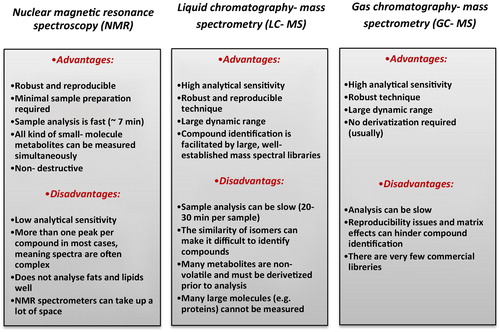
NMR is considered as one of the most commonly used analytical platforms in metabolomics. Single proton 1H NMR involves the use of a large and powerful magnet to align the protons which are available in a sample. In this method, each metabolite appears with a distinct peak in the NMR spectrum and the area under the peak represents each metabolite concentration. The advantages and disadvantages of this method are summarized in . One of the most important advantages of NMR for detecting metabolites in this method is that the sample is not degraded and it is even used in subsequent studies.Citation3 Low sensitivity and detection of limited mass range are regarded as the limitation of NMR. The general process of NMR-based metabolomics includes the following stepsCitation16
| • | Experimental design | ||||
| • | Sample preparation | ||||
| • | NMR raw data | ||||
| • | Spectral processing | ||||
| • | Data preprocessing | ||||
| • | Multivariate data analysis (PCA, PLS-DA, OPLS-DA) | ||||
| • | Metabolic pathway identification (potential biomarkers) | ||||
3.1.2 MS-based techniques
Metabolite spectrum data are considered as mass-to-charge (m/z) ratios by MS.Citation24 MS is primarily coupled with liquid chromatography or gas chromatography in metabolomics studies.Citation3 It is worth noting that the use of LC-MS in metabolomics studies results in detecting a wide range of metabolites.Citation25 Regarding some idiosyncratic properties of LC-MS method, we can refer to the simultaneous assessment of aqueous and lipid metabolites, more separation, and detection of different types of metabolites.Citation26 This method covers a greater mass range and no derivation of samples is required. However, the variability of LC-MS instruments and absence of standard metabolic library are considered as the disadvantages of this approach .Citation27 In order to detect metabolites by LC-MS, some steps should be taken such as separating by chromatography, ionizing the sample, determining the compound mass by mass analyzer, and detecting the metabolites by detector. TOF and quadrupole are regarded as the most common mass analyzers coupled with LC. During recent years, some advance techniques such as FT-ICR with high mass accuracy and consecutive MS (tandem MS) ability has attracted a lot of attention. However, it is less implemented due to the high cost of FT-ICR. GC is another separation method which is coupled with MS. GC-MS is used for separating and detecting volatile metabolites at high temperature with high sensitivity. The efficiency of this method is related to the high sensitivity and the detection of volatile metabolites capability. However, regarding the major challenges of GC-MS, we can refer to the derivation of readily volatile samples and limited mass range that incomplete derivation can lead to loss of metabolite and intricate explanation of results.Citation26
3.2 Statistical analysis of metabolomics-based data
Multi-dimensional dataset includes the signal from a large number of metabolites generated in metabolomics studies. Thus, developing a coherent hypothesis explaining any changes in metabolite profile by intuition alone is difficult due to a large number of measurements. Therefore, employing a multi-dimensional statistical analysis is essential for reducing the number of variables in a dataset. The most common methods used for reducing data are related to unsupervised methods such as PCA and supervised methods including PLS, PLS-DA.Citation28 PCA method is utilized for outlier identification within the dataset. Then, PLS-DA and OPLS-DA as unsupervised multivariate data analytical methods are performed for 1H NMR spectral data analysis. PLS-DA is considered as a pattern recognition approach used to identify potential disease-specific biomarkers. After identifying the metabolites, their interpretation into biological process and pathways is an important step in analyzing the results of metabolomics study.Citation29 The existence of valid database including invaluable information about metabolites and the involved pathways play a significant role in mapping metabolites on biological system. There are a number of public information databases about metabolites and their reactions, as well as some of open source tools such as MetaCore which provide some helpful ways to experimentally map the observed changes of metabolites into metabolic pathways.Citation3,Citation30 Finally, the data are imported into the Metscape plugin by Cytoscape software for more analysis of differentiated metabolites in the groups under study. Metscape plugin use to map of the pathway of metabolomics data. Finally, this application demonstrates seed metabolites, metabolic reactions, enzymes, and genes.Citation3,Citation31
4 Metabolomics and Leishmania/leishmaniasis
Recently, a number of metabolomics-based studies have been published with respect to Leishmania/leishmaniasis. Metabolomics as a new field in post-genomic era represents an emerging discipline related to a comprehensive analysis of small molecule metabolites, which provides a powerful approach to discover biomarkers in biological systems. L. major Fredlin strain is the first protozoan model which metabolic network was reconstructed by Chavali et al.Citation32 In this regard, NMR and MS spectrometry are considered as the most useful methods.
4.1 1H NMR-based metabolomics in the leishmania/leishmaniasis research
Most of the studies in the field of Leishmania by 1HNMR-based metabolomics have been conducted on life cycle stages in metabolic changes of parasite in different species. However, metabolomics-based studies about the drugs affected metabolomics map have been less emphasized and only one study was reported on blood serum of cutaneous leishmaniasis. indicates all of the previous studies on Leishmania by 1H NMR-based metabolomics. Arjmand et al.Citation33 focused on metabolomics by using 1H NMR in order to compare two stages of promastigote forms of parasite including procyclic and metacyclic in L. major species. The main altered metabolites in logarithmic and stationary phases included citraconic acid, isopropylmalic acid, L-leucine, ornithine, caprylic acid, capric acid, and acetic acid. Amino acids play an important role in energy production in Leishmania parasite.Citation34 The major alterations in amino acids in metacyclogenesis process include citraconic acid, isopropylmalic acid, L-leucine related to valine, leucine and isoleucine biosynthesis pathways, respectively. In the present study, caprylic and capric acids in fatty acid biosynthesis pathway indicated some alterations between the two phases. Based on the results of the previous studies, the oxidation rate in fatty acids is faster in stationary phase rather than the logarithmic phase, which is consistent with the results of previous studies.Citation35 The final significant change in metabolites is related to acetic acid in pyruvate and sulfur metabolic pathways. In the mentioned study, the detection of metabolite alterations could be helpful for better understanding of metacyclogenesis process and reopening new horizons in control of disease. In another study, Boroujeni et al.,Citation36 performed NMR-based approach for metabolic profiling of L. major in the control group (without artemisinin reception in culture media) and the case group (with artemisinin reception in culture media). Based on the results, the most significant changes were related to galactose metabolic cycle. Other significant metabolites belong to sphingolipid, valine, leucine and isoleucine biosynthesis pathways. The results showed that galactose metabolism is the most important pathway in cutaneous leishmaniasis pathogenesis and can be considered as an important drug target in parasite extermination. According to sphingolipid role in apoptosis, the results of this study on the changes of this metabolic pathway indicate that artemisinin is effective for parasite clearance. Drug resistance represents a major challenge for controlling leishmanial infectious. In addition, based on the results, antimonial (sodium stibogluconate; SSG) resistance was closely related to metacyclogenesis process in L. donovani as SSG-resistant strains could increase the capacity for generating metacyclic-infective forms. Also, by using 1H NMR, Lamour et al.,Citation37 characterized the cell growth medium, parasite extract and intra and extracellular metabolome of activated and non-activated macrophages in the presence and absence of L. major. The results demonstrated some metabolic differences between infection state and macrophage activation. Succinate, acetate, and alanine metabolites placed at higher level in the samples exposed to parasite, compared to uninfected macrophages. Furthermore, Gupta et al.,Citation38 characterized map/profile of in vitro cultured axenic amastigote metabolites, compared to intracellular amastigote and promastigote forms employing proton NMR spectroscopy. In this study, two new metabolites including betaine and β-hydroxybutyrate were reported. It is worth noting that betaine was presented in all of parasite forms while β-hydroxybutyrate was identified only in the promastigote and axenic amastigote stages. Among other metabolites, acetoacetate could only be detected in axenic and intracellular amastigote stages. All the remaining metabolites were present at all three stages of parasite which were considered in the present plan with different levels. Tafazzoli et al.,Citation39 in another study, utilized a non-targeted study by using gas NMR in order to compare the metabolite profiling of 20 leishmaniasis patients sera and 44 healthy individuals. In addition, they measured the concentrations of selenium in all 64 serum samples by using an atomic absorption device. In this study, PCA and PLS methods were applied for distinguishing the outliers and classifying the patients based on selenium concentration in blood serum. The most important metabolites were related to threonine, lipids, lactate and alanine. The findings of this study indicated a significant relationship between selenium concentration and the related variables. In case of drug resistance mechanism evaluation in leishmania, Rainey and Mackenzie, 1991 performed metabolomics study in order to clarify the drug resistance mechanisms in Leishmania species which is regarded as a problem in treatment output.Citation40
Table 1 Summary of 1H NMR-based metabolomics studies conducted in the field of leishmania research.
4.2 MS-based metabolomics in the leishmania/leishmaniasis research
Metabolomics studies based on mass spectrometry are proving of great utility in the analysis of different aspects of protozoan parasites such as leishmania genus. Metabolomics has also given insight into the biology, pathogenesis, mechanisms of drug against parasites and also drug resistance in leishmania. The list of LC-MS-based metabolomics studies along with details of the studies are summarized in . In a metabolomics investigation, Akpunarlieva et al.,Citation41 integrated proteomics and metabolomics approaches to identify proteins and metabolites, which represent altered abundance in mutant L. Mexicana (ΔLmGT) promastigotes. In this study, the molecular variations of wild-type and mutant model of Leishmania were investigated in proteome and metabolome levels. The results indicated some variations such as deficiency in glucose transport and several phenotypic changes in mutant model, as well as seeking molecular interpretation of these phenotypes. In addition, the results demonstrated a series of changes in metabolic pathways of glycoconjugate production and redox homeostasis involved in glucose metabolism in response to oxidative stress, which may represent the compatibility to the loss of sugar uptake capacity and elucidate the low virulence of this mutant among the hosts. In other study, Westrop et al.,Citation42 compared the metabolome profile of promastigotes of L. donovani, L. major and L. mexicana by LC-MS technique. The analysis revealed 64 metabolites with confirmed identity differing 3-fold or more between the cell extracts of species and 43 metabolites of confirmed identity in culture media of species, respectively. Furthermore, the recognized metabolites were detected in cell extracts of three species and their culture media included 161 and 87 metabolites. Significant differences in metabolic pathways were observed in amino acids, especially Trp, Asp, Arg and Pro, compared to the global metabolome of the studied Leishmania spp. This study failed to play a role in detecting inter-species differences about metabolic adaptation of parasites in their hosts. In this regard, Vincent et al.,Citation43 implemented an untargeted LC-MS metabolomics approach to clarify the metabolic profile changes involved in miltefosine action at time intervals of 0, 1.25, 2.5, 3.75 and 5 h of drug treatment in L. infantum species. Totally, 876 metabolites were detected, among which alkane fragment and sugar release related to metabolites significantly altered after 3.75 h. Fragment release was concurrent with ROS production, while no changes has occurred in membrane phospholipids. In addition, the levels of some thiols and polyamines changed indicating DNA damage in the treated cells. Due to cell membrane damage, the evacuated levels of most metabolites occurred after 5 h of miltefosine treatment. In miltefosine resistant cells, no change was found in the internal metabolites levels. The results are important for understanding miltefosine and can be regarded as fundamental information in designing treatment program. In this respect, Berg et al.,Citation44 implemented an untargeted LC-MS platform for evaluating the metabolic differences of L. donovani during promastigote stages in the absence or presence of drug pressure. The study presented new approaches in the context of antimonial resistance and differentiation. The results of metabolites changes demonstrated that the SSG-resistant parasites can play a significant role in encountering with oxidative stress process. Further, Silva et al.,Citation45 applied an untargeted metabolomics approach for evaluating the metabolic changes during promastigote by developing in vitro culturing between 3 and 6 days. Based on the results, the metabolome profile of promastigotes differed significantly from each other during culture. The most striking metabolic differences were related to the structural variations in glycerophospholipids and an increase in the amount of sphingolipids and glycerolipids as cells progressed from procyclic to metacyclic stages. Furthermore, Kindt et al.,Citation46 implemented an untargeted LC-MS metabolomics approach to global comparative metabolic profiling of antimonial-sensitive and antimonial-resistant L. donovani isolates. They determined the antimonial-sensitive and antimonial-resistant differences by using metabolomics study and measured the relative abundance of 340 metabolites and a large number of lipids. Based on the results, distinct metabolic profiles were observed in drug-sensitive and drug-resistant parasites and significant differences were shown between two phenotypes among100 metabolites. The most significant difference in metabolites was related to the pathways of lipid metabolism. However, the results failed to involve resistance mechanisms for antimonial drugs. Also, in the field of comparative metabolomics of wild type and mutant parasite model, De Souza et al.,Citation47 described a GC-MS peak clustering approach for discriminating the wild-type of L. Mexicana from two mutant parasite lines based on their metabolic profile.
Table2 Summary of MS-based metabolomics studies conducted in the field of leishmania research.
5 Conclusion
Leishmania as a protozoan parasite is considered as an etiological agent of wide range of clinical manifestations known as “leishmaniasis”. The disease represents a major public health problem and is endemic in 98 countries across five continents. However, no effective vaccine and decisive treatment has been introduced yet. Over the past few years, “Omics” technologies have played a pivotal role in recognizing parasite biology in its different life cycle stages, identifying the parasite-host interaction, and introducing novel drug target. Metabolomics approach based on a large-scale study of metabolite profiles both in parasite and the host can accelerate the discovery of novel metabolite markers and reveal their related biochemical pathways to understand the underlying pathology of leishmaniasis. In addition, metabolomics approach has attracted a lot of attention among researchers, due to the regulation of gene expression of trypanosomatidae in post-transcriptional level. This review provides a reference including all studies in the field of Leishmania metabolomics. In most of the conducted studies, metabolomics-based study of L. donovani, L. major and L. mexicana spp. was investigated by LC-MS and NMR methods while other species such as L. tropica, L. infantun, etc. were neglected. The most common objectives in these studies are related to a comparative metabolomics of the biology of Leishmania parasite in different stages such as procyclic, metacyclic and amastigote. However, a few studies were conducted by using biological samples of mammalians hosts for example blood, urine and tissue samples in response to infection. In this respect, the results of the study on blood serum of cutaneous leishmaniasis showed that the most important metabolites include threonine, lipid, lactate and alanine. Further, the limitation of metabolomics study of parasite isolates has great potential to increase our understanding of metabolism in the clinically relevant stage. The study of drug resistance and drug action by metabolomics obtain some important information about designing integrated therapies, understanding parasite metabolism, and highlighting some new targets for chemotherapy important. In metabolomics-based study, the study of metabolic pathways in natural plant-derived products is very critical in confronting the disease in a natural program with the least amount of side effects. The present review will provide a reference to conduct new studies for better understanding of the pathways playing a role in the biology of Leishmania and its control. Additionally, the integration of metabolomics with other platforms of system biology such as genomics, transcriptomics, proteomics and lipidomics will provide further and better insights into evaluating different pathophysiology aspects and treating the disease.
Conflict of interest
The authors declare that there is no conflict of interest.
Acknowledgements
The authors are thankful to the Proteomics Research Center and Vice-Chancellor for Research in Shahid Beheshti University of Medical Sciences, Iran, for their support.
Notes
Peer review under responsibility of Alexandria University Faculty of Medicine.
Available online 2 July 2018
References
- N.A.D.AtanM.KoushkiM.R.TaviraniN.A.AhmadiProtein-protein interaction network analysis of salivary proteomic data in oral cancer casesAsian Pac J Cancer Prv19201816391645
- F.S.SadjjadiM.Rezaie-TaviraniN.A.AhmadiS.M.SadjjadiH.ZaliProteome evaluation of human cystic echinococcosis sera using two dimensional gel electrophoresisGastroenterol Hepatol Bed Bench1120187582
- K.A.StringerR.T.McKayA.KarnovskyB.QuémeraisP.LacyMetabolomics and its application to acute lung diseasesFront Immunol7201622 p
- M.Rezaei-TaviraniF.FathiF.DarvizehAdvantage of applying OSC to 1H NMR-based metabonomic data of celiac diseaseInt J Endocrinol Metab102012548552
- A.SafaeiA.A.OskouieS.R.MohebbiMetabolomic analysis of human cirrhosis, hepatocellular carcinoma, non-alcoholic fatty liver disease and non-alcoholic steatohepatitis diseasesGastroenterol Hepatol Bed Bench92016158173
- G.J.PattiO.YanesG.SiuzdakInnovation: metabolomics: the apogee of the omics trilogyNat Rev Mol Cell Biol132012263269
- D.S.WishartComputational approaches to metabolomics. Bioinformatics methods in clinical researchMethods Mol Biol2010283313
- J.XiaD.I.BroadhurstM.WilsonD.S.WishartTranslational biomarker discovery in clinical metabolomics: an introductory tutorialMetabolomics92013280299
- N.A.AhmadiM.ModiriS.MamdohiFirst survey of cutaneous leishmaniasis in Borujerd county, western Islamic Republic of IranEast Mediterr Health J192013847853
- J.-C.DujardinStructure, dynamics and function of Leishmania genome: resolving the puzzle of infection, genetics and evolution?Infect Genet Evol92009290297
- World Health OrganizationReport of the consultative meeting on cutaneous leishmaniasis2008WHOGeneva31
- S.KumariA.KumarM.SamantN.SinghA.DubeDiscovery of novel vaccine candidates and drug targets against visceral leishmaniasis using proteomics and transcriptomicsCurr Drug Targets92008938947
- S.SundarDrug resistance in Indian visceral leishmaniasisTM & IH62001849854
- S.GoyardH.SegawaJ.GordonAn in vitro system for developmental and genetic studies of Leishmania donovani phosphoglycansMol Biochem Parasitol13020033142
- F.McNicollJ.DrummelsmithM.MüllerA combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantumProteomics6200635673581
- L.ZhangE.HatzakisA.D.PattersonNMR-based metabolomics and its application in drug metabolism and cancer researchCurr Pharmacol Rep22016231240
- A.M.SliszA.P.BreksaIIID.O.MishchukG.McCollumC.M.SlupskyMetabolomic analysis of citrus infection by ‘Candidatus Liberibacter’ reveals insight into pathogenicityJ Proteome Res11201242234230
- N.L.KuehnbaumP.Britz-McKibbinNew advances in separation science for metabolomics: resolving chemical diversity in a post-genomic eraChem Rev113201324372468
- B.F.NobakhtR.AliannejadM.Rezaei-TaviraniS.TaheriA.A.OskouieThe metabolomics of airway diseases, including COPD, asthma and cystic fibrosisBiomarkers202015516
- Ah.ZhangH.SunS.QiuX.J.WangNMR-based metabolomics coupled with pattern recognition methods in biomarker discovery and disease diagnosisMagn Reson Chem5120135491456
- J.K.NicholsonJ.C.LindonSystems biology: metabonomicsNature455200810541056
- M.ScholzS.GatzekA.SterlingO.FiehnJ.SelbigMetabolite fingerprinting: detecting biological features by independent component analysisBioinformatics20200424472454
- B.F.NobakhtR.AliannejadM.Rezaei-TaviraniNMR-and GC/MS-based metabolomics of sulfur mustard exposed individuals: a pilot studyBiomarkers212016479489
- A.AlonsoS.MarsalA.JuliàAnalytical methods in untargeted metabolomics: state of the art in 2015Front Bioeng Biotechnol32015120
- H.G.GikaG.A.TheodoridisR.S.PlumbI.D.WilsonCurrent practice of liquid chromatography–mass spectrometry in metabolomics and metabonomicsJ Pharm Biomed Anal8720141225
- K.M.SasA.KarnovskyG.MichailidisS.PennathurMetabolomics and diabetes: analytical and computational approachesDiabetes642015718732
- N.J.SerkovaT.J.StandifordK.A.StringerThe emerging field of quantitative blood metabolomics for biomarker discovery in critical illnessesAm J Respir Crit Care Med1842011647655
- O.A.JonesMetabolomics and systems biology in human health and medicine2014Cabi
- ZhangA-hH.SunYanG-lY.YuanY.HanX-j.WangMetabolomics study of type 2 diabetes using ultra-performance LC-ESI/quadrupole-TOF high-definition MS coupled with pattern recognition methodsJ Physiol Biochem702014117128
- F.García-AlcaldeF.García-LópezJ.DopazoA.ConesaPaintomics: a web based tool for the joint visualization of transcriptomics and metabolomics dataBioinformatics272010137139
- A.KarnovskyT.WeymouthT.HullMetscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression dataBioinformatics282011373380
- A.K.ChavaliJ.D.WhittemoreJ.A.EddyK.T.WilliamsJ.A.PapinSystems analysis of metabolism in the pathogenic trypanosomatid Leishmania majorMol Syst Biol42008177
- M.ArjmandA.MadrakianG.KhaliliA.NajafiZ.ZamaniZ.AkbariMetabolomics-based study of logarithmic and stationary phases of promastigotes in Leishmania major by 1H NMR spectroscopyIran Biomed J2020167783
- F.R.OpperdoesG.H.CoombsMetabolism of Leishmania: proven and predictedTrends Parasitol232007149158
- J.BlumEffects of culture age and hexoses on fatty acid oxidation by Leishmania majorJ Eukaryot Microbiol371990505510
- M.Beigi BoroujeniM.ArjmandG.KhaliliZ.AkbariA.NajafiA.ShafieiThe metabonomic changes of leishmania major, s promastigotes (fredlin strain) after in vitro artemisinin treatment at stationary phaseKoomesh1620149096
- S.D.LamourB.-S.ChoiH.C.KeunI.MüllerJ.SaricMetabolic characterization of Leishmania major infection in activated and nonactivated macrophagesJ Proteome Res11201242114222
- N.GuptaN.GoyalU.SinghaV.BhakuniR.RoyA.RastogiCharacterization of intracellular metabolites of axenic amastigotes of Leishmania donovani by 1H NMR spectroscopyActa Tropica731999121133
- M.TafazzoliF.FathiF.DarvizehZ.ZamaniF.PourfallahA metabonomic study on samples of cutaneous leishmaniasis and its correlation with the selenium level in the blood serumJ Mol Biomark Diagn12010101
- P.M.RaineyN.E.MacKenzieA carbon-13 nuclear magnetic resonance analysis of the products of glucose metabolism in Leishmania pifanoi amastigotes and promastigotesMol Biochem Parasitol451991307315
- S.AkpunarlievaS.WeidtD.LamasudinIntegration of proteomics and metabolomics to elucidate metabolic adaptation in LeishmaniaJ Proteomics15520178598
- G.D.WestropR.A.WilliamsL.WangMetabolomic analyses of Leishmania reveal multiple species differences and large differences in amino acid metabolismPloS One102015e0136891
- I.M.VincentS.WeidtL.RivasK.BurgessT.K.SmithM.OuelletteUntargeted metabolomic analysis of miltefosine action in Leishmania infantum reveals changes to the internal lipid metabolismInt J Parasitol Drugs Drug Resist420142027
- M.BergM.VanaerschotA.JankevicsMetabolic adaptations of Leishmania donovani in relation to differentiation, drug resistance, and drug pressureMol Microbiol902013428442
- A.M.SilvaA.Cordeiro-da-SilvaG.H.CoombsMetabolic variation during development in culture of Leishmania donovani promastigotesPLoS Negl Trop Dis52011e1451
- R.t’KindtA.JankevicsR.A.ScheltemaTowards an unbiased metabolic profiling of protozoan parasites: optimisation of a Leishmania sampling protocol for HILIC-orbitrap analysisAnal Bioanal Chem398201020592069
- D.P.De SouzaE.C.SaundersM.J.McConvilleV.A.LikićProgressive peak clustering in GC-MS metabolomic experiments applied to Leishmania parasitesBioinformatics22200613911396