Protein microarray analysis of apoptosis-related protein expression following heat shock in human tongue squamous cell carcinomas containing different p53 phenotypes. Int. J. Hyperthermia, 2008; 24(8), 605–612.
International Journal of Hyperthermia hereby notifies its readers that errors were published in the article listed above. The corrections are shown in bold face below:
p. 608, right column, 3rd paragraph:
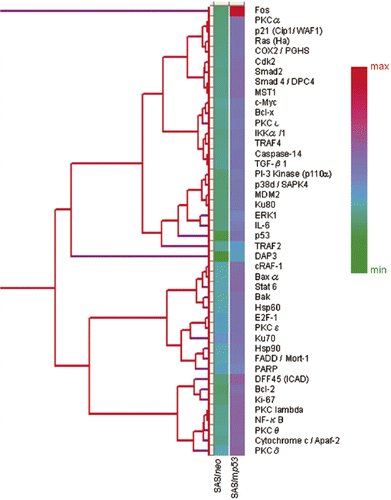
In Table III, among the proteins which inhibit apoptosis, Bcl-2 (B–cell lymphoma/leukemia-2), COX (cyclooxygenase)-2, PKCα, PKCβ, DFF45 (DNA fragmentation factor-45), PKCθ, Bcl-xL (B-cell CLL/lymphoma × the long isoform), MDM2 (murine double minute 2), NF-κB (nuclear factor κB), Rb2 (retinoblastoma protein 2), STAT3 (signal transducer and activator of transcriptional 3), TRAF2 (tumor necrosis factor receptor associated factor 5), IL-6 (interleukin 6), PKCλ, PKCτ and IKK-α/1 (I-κ-B kinase-α/1) were up-regulated in SAS/mp53 cells. FADD/Mort-1, Ku70, PKCδ, PKCε and E2F-1 were up-regulated in SAS/mp53 cells as well as in SAS/neo cells. Other apoptosis-inducing proteins were also up-regulated in SAS/mp53 cells. These were Baxα (Bcl-2 associated × protein α), cRAF-1 (v-raf-1 murine leukemia vial oncogene homolog 1), TGF-β1(transforming growth factor β1), cytochrome c/Apaf-2 (apoptotic protease activating factor 2), c-Myc (cellular homologue of avian myelocytomatosis virus oncogene), Bak (Bcl-2 homologous antagonist/killer), DAP3 (death-associated protein kinase 3), Smad2 (Sma- and Mad-related protein 2) and Smad4/DPC4 (Sma- and Mad-related protein 4/deleted in pancreatic cancer locus 4). The level of induced Hsp90 (heat shock protein 90) in SAS/mp53 cells was higher than that induced in SAS/neo cells.
p. 611, left column, 1st paragraph:
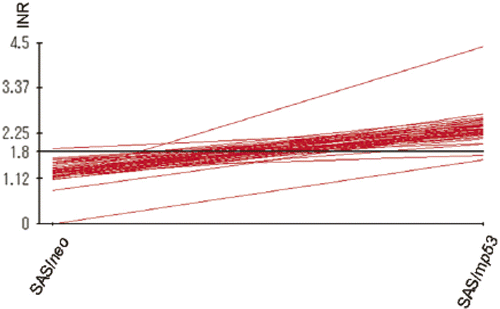
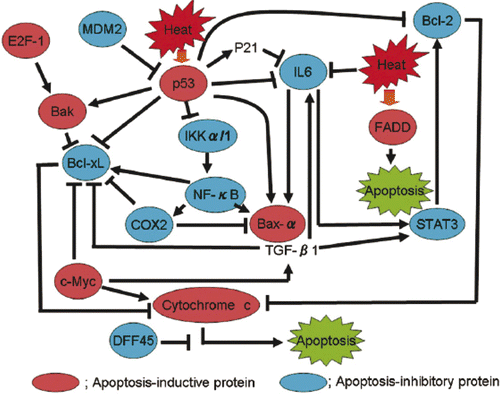
The results reported here show that apoptosis-inhibitory proteins (such as Bcl-2, Bcl-xL, IL-6, and others) were strongly induced by heat treatment in SAS/mp53 cells when compared to SAS/neo cells containing a wtp53.
Addendum
Color plates for through 4 are found on the following page for the print edition.
Figure 1. Unsupervised hierarchical clustering analysis of 512 proteins in heat treated cells compared with non-treated cells. The graph indicates relative protein expression levels. Lower signal values are indicated by green, and higher signal values are indicated by red.
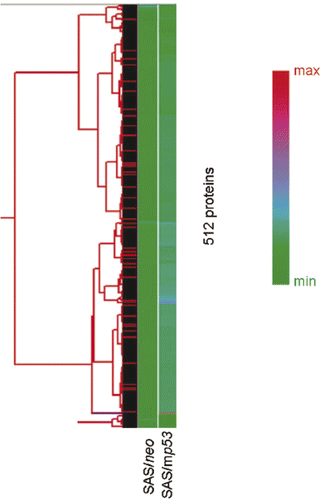
Figure 2. Hierarchical clustering analysis of 44 selected apoptosis-related proteins which showed valid changes in heat treated cells when compared to non-treated cells. The graph indicates protein expression levels with lower signal values shown in green and higher signal values shown in red.