Abstract
Background:
The pathogenesis of pulmonary fibrosis is not fully understood. Previous work has demonstrated the important role of circular RNA (circRNA) in pulmonary fibrosis development. This study aims to analyse the role of circ_0035796 in pulmonary fibrosis and the underlying mechanism.
Methods:
Human foetal lung fibroblast 1 (HFL1) cells were treated with transforming growth factor-β1 (TGF-β1) to mimic a pulmonary fibrosis cell model. The expression of circ_0035796, microRNA-150-5p (miR-150-5p) and L1 cell adhesion molecule (L1CAM) was determined by quantitative real-time polymerase chain reaction (qRT-PCR). The protein expression of L1CAM, collagen I and fibronectin was detected by Western blot. Cell viability was analysed by CCK-8 assay. Cell proliferation, invasion and migration were investigated by 5-Ethynyl-2’-deoxyuridine (EdU) assay, transwell invasion assay and wound-healing assay, respectively. The secretion of interleukin-6 (IL-6) and tumour necrosis factor-α (TNF-α) was analysed by Enzyme-linked immunosorbent assay (ELISA). Oxidative stress was assessed by detecting Superoxide Dismutase (SOD) activity and Malondialdehyde (MDA) level using commercial kits. The association of miR-150-5p with circ_0035796 and L1CAM was identified by dual-luciferase reporter assay, RNA pull-down assay and RNA immunoprecipitation (RIP) assay.
Results:
Circ_0035796 and L1CAM expression were dramatically upregulated, while miR-150-5p expression was downregulated in TGF-β1-treated HFL1 cells. TGF-β1 treatment induced cell proliferation, migration, invasion, IL-6 and TNF-α secretion, and oxidative stress, whereas circ_0035796 depletion relieved these effects. In addition, circ_0035796 acted as a sponge of miR-150-5p and miR-150-5p combined with L1CAM. Moreover, miR-150-5p depletion attenuated circ_0035796 knockdown-mediated effects in TGF-β1-exposed HFL1 cells. The regulation of miR-150-5p on TGF-β1-induced fibroblast activation involved the downregulation of L1CAM. Further, circ_0035796 modulated L1CAM expression by interacting with miR-150-5p in TGF-β1-exposed HFL1 cells.
Conclusion:
Circ_0035796 knockdown ameliorates TGF-β1-induced pulmonary fibrosis through the miR-150-5p/L1CAM axis in vitro.
1. Introduction
Pulmonary fibrosis is a chronic lung disease, featured by inappropriate scar tissue formation and persistent alveolar epithelial injury in the lung [Citation1]. The potential triggers of pulmonary fibrosis include inhaled toxic material, smoking, infection, ionising radiation, and chemotherapy [Citation2]. The pathogenesis of the disease involves abnormal tissue remodelling, immune response, cell apoptosis, coagulation, fibroblast-to-myofibroblast transition and extracellular matrix (ECM) deposition [Citation3,Citation4]. In terms of mechanism, the dysregulation of profibrotic and antifibrotic mediators, such as transforming growth factor-β (TGF-β), leads to defective regeneration as well as aberrant remodelling, finally resulting in the occurrence of pulmonary fibrosis [Citation4]. Thus, investigation of the detailed mechanism of pulmonary fibrosis may provide more effective therapeutic options for the disease.
CircRNA is a special endogenous RNA and has no 5'-3’ polarity, showing dynamic expression patterns in various diseases [Citation5]. The unique circular structure of circRNAs endows a stable status to this kind of RNA in cells [Citation6]. Most circRNAs are expressed in the cytoplasm and have many miRNA response elements, therefore competitively binding to microRNA (miRNA) [Citation7]. Currently, the endogenous competitive roles of circRNAs to miRNAs have been widely reported in multiple diseases, including pulmonary fibrosis [Citation7,Citation8]. A recent study showed that circHIPK3 increased glycolysis through miR-30a-3p, thus facilitating fibroblast activation [Citation9]. Circ_0044226 depletion repressed pulmonary fibrosis development through the regulation of cell differentiation cyclin 27 (CDC27) [Citation10]. Another circRNA, circ_0035796, is significantly upregulated in pulmonary fibrosis patients, as determined by hierarchical clustering according to its fold change in the pulmonary fibrosis group and the normal group [Citation11]. However, whether the pathogenesis of pulmonary fibrosis involves circ_0035796 remains unknown.
MiRNA emerges as a small RNA and regulates gene expression through transcript degradation and/or translation repression during different biological processes [Citation12]. Considerable research demonstrates that miRNAs affect the development of different diseases, such as pulmonary and cardiovascular diseases [Citation13,Citation14]. Some miRNAs like miR-21 [Citation15] and miR-410 [Citation16] are abnormally expressed in fibrotic conditions and may participate in fibrotic activation. MiR-150-5p is an extensively studied miRNA and plays important role in the occurrence of pulmonary diseases [Citation17,Citation18]. Through prediction of the starbase database, we found that circ_0035796 potentially bound to miR-150-5p. Given that fibroblast-to-myofibroblast transition involved miR-150-5p [Citation19], we hypothesised that the regulation of circ_0035796 towards pulmonary fibrosis development involved miR-150-5p, while no investigator performed the study on the role of the circ_0035796/miR-150-5p axis in pulmonary fibrosis.
Thus, in this study, we analysed the role of circ_0035796 in the pulmonary fibrosis process including cell proliferation, invasion, migration, inflammation and oxidative stress using TGF-β1-induced human foetal lung fibroblast 1 (HFL1) cells and determined whether circ_0035796 regulated pulmonary fibrosis process by targeting miR-150-5p and regulating L1 cell adhesion molecule (L1CAM).
2. Materials and methods
2.1. Cell culture and treatment
HFL1 cells were purchased from Y-J Biotechnology Co., Ltd. (Shanghai, China) and cultured in RPMI-1640 (Biosun, Shanghai, China) plus 10% foetal bovine serum and 1% penicillin/streptomycin at 37 °C with 5% CO2. To generate a pulmonary fibrosis cell model, HFL1 cells were induced using 6 ng/mL TGF-β1 (Thermo Fisher, Waltham, MA, USA) for 48 h. The cells treated with the same amount of RPMI-1640 served as controls.
2.2. Cell transfection
Songon Biotech (Shanghai, China) provided the small interfering RNAs of circ_0035796 (si-circ_0035796, 5'-GCATC ACTATCAGATATCAGA-3') and L1CAM (si-L1CAM,5'-CATCTACCGCTGCTTTGCCAGCAAT-3'), the mimics of miR-150-5p (5'-UCUCCCAACCCUUGUACCAGUG-3'), the inhibitors of miR-150-5p (anti-miR-150-5p, 5'-CACUGG UACAAGGGUUGGGAGA-3'), and the matched controls (si-NC, si-con, miR-NC, and anti-miR-NC). L1CAM overexpression plasmid was achieved by introducing the coding sequence of L1CAM into the pcDNA 3.1 vector. Based on the manufacturer’s direction of FuGENE6 (Roche, Basel, Switzerland), HFL1 cells were seeded in 12-well plates and cultured for 12-18 h, followed by transfection for 48 h.
2.3. Quantitative real-time polymerase chain reaction (qRT-PCR)
We separated nuclear/cytoplasmic fractionations of HFL1 cells using ARIS Kit (Thermo Fisher). Total RNA was isolated using RNAprep Pure Tissue reagents (Tiangen, Beijing, China). For RNase R treatment assay, 2 μg RNA isolated from HFL1 cells was incubated with RNase R (Geneseed, Guangzhou, China) for 20 min. Reverse transcription reaction was conducted using FastKing RT Kit (Tiangen) and a specific miRNA synthesis kit (Thermo Fisher) as per the guidebooks. Subsequently, SYBR Green Mix (Tiangen) was used for qRT-PCR analysis. Data were processed by the 2-ΔΔCt method with the normalisation to U6 or GAPDH. Primer sequences are listed in . Random primers and oligo(dT)18 primers were used to identify the circular characteristic of circRNA.
Table 1. Primer sequences for qRT-PCR.
2.4. Cell counting kit-8 (CCK-8) assay
HFL1 cells were seeded in 96-well plates overnight and treated with various reagents, followed by culturing for 48 h. Then, CCK-8 reagent (Beyotime, Shanghai, China) was used to culture the cells, lasting 3 h. Finally, microplate reader (REAGEN, Shenzhen, China) was utilised to detect samples.
2.5. 5-Ethynyl-2’-deoxyuridine assay
After 48 h of treatment, plates were added with 5-Ethynyl-2’-deoxyuridine (EdU) labelling RPMI-1640 and further incubated for 2 h at 37 °C according to the guidebook of EdU detection reagents (Ribobio, Guangzhou, China). Subsequently, the cells were incubated with Triton X-100, paraformaldehyde and anti-EdU working solution. Finally, EdU-positive cells were determined under fluorescent microscopy.
2.6. Transwell invasion assay
In terms of cell invasion analysis, we employed 24-well transwell compartments, which were coated with Matrigel (Qcbio science, Shanghai, China) in advance. HFL1 cells suspended in serum-free RPMI-1640 were added into the upper chambers. After a 24-h incubation, HFL1 cells on the upper chambers were removed, and methanol was added to the lower membrane surface. Finally, a microscope was used for cell counting.
2.7. Wound-healing assay
After various treatments, 5 × 105 HFL1 cells were seeded in each well of 6-well plates and cultured to 30–50% confluence. Then, the production of confluent cellular monolayer was conducted using 10 μL pipette tips. These cells were cultured with RPMI-1640 plus 1% foetal bovine serum. Twenty-four hours later, a microscope was used to capture images.
2.8. Western blot analysis
Total proteins from HFL1 cells were extracted following the users’ instructions for RIPA lysis buffer (Beyotime). 20 μg protein, as determined using NanoDrop 2000 spectrophotometer (Thermo Fisher), was separated by polyacrylamide gels. Membranes were blocked with defatted dry milk prior to incubating with primary antibodies targeting collagen I (Cat#PA1-26204; 1:1000; Thermo Fisher), fibronectin (Cat#PA1-23693; 1:1000; Thermo Fisher), L1CAM (Cat#PA5-85876; 1:1000; Thermo Fisher), and GAPDH (Cat#437000; 1:500; Thermo Fisher). Then, the membranes were probed with secondary antibodies (Thermo Fisher) and immediately imaged using BioSpectrum Imaging System. Protein expression was analysed using Image J software with normalisation to GAPDH.
2.9. Enzyme-linked immunosorbent assay
The levels of interleukin-6 (IL-6) and TNF-α in HFL1 cell supernatant after appropriate transfections and/or treatments were assessed with commercial Enzyme-linked immunosorbent assay (ELISA) kits (Beyotime) referring to the guidebooks. Microplate reader (REAGEN) was used to detect these samples.
2.10. Determination of malondialdehyde (MDA) level and Superoxide dismutase (SOD) activity
After appropriate transfections and/or treatments, 2 × 106 HFL1 cells were harvested and analysed following the instructions of MDA assay kit (ab118970; Abcam, Cambridge, MA, USA) and SOD activity assay kit (ab65354; Abcam). Measurement of absorbance was performed on a microplate reader (REAGEN).
2.11. Dual-luciferase reporter assay
After prediction of the binding sites of miR-150-5p and circ_0035796 or L1CAM through the starbase online database (https://rnasysu.com/encori/), the wild-type (WT) and mutant (MUT) reporter plasmids of circ_0035796 and L1CAM were generated by Songon Biotech, and named as WT-circ_0035796, MUT-circ_0035796, WT-L1CAM 3’UTR and MUT-L1CAM 3’UTR. Then, HFL1 cells were co-transfected with the above reporter plasmids, miR-150-5p mimics or miR-NC for 48 h, referring to the guidebook of FuGENE6 (Roche). At last, Dual-Lucy Assay Kit (Solarbio, Beijing, China) was employed for the detection of luciferase activity.
2.12. RNA pull-down assay
GenePharma Co., Ltd. (Shanghai, China) designed biotin-labeled miR-150-5p (bio-miR-150-5p) and bio-miR-NC for RNA pull-down assay. Cell lysates extracted from HFL1 cells have undergone two-hour incubation with bio-miR-150-5p and bio-miR-NC and Streptavidin-coupled Dynabeads (Sigma, St. Louis, MO, USA) were employed for capturing the miR-150-5p-associated miRNA and mRNA complexes. Eventually, qRT-PCR analysis of the pull-down RNAs was conducted.
2.13. RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assay implicating the use of Magna RNA immunoprecipitation kit (Millipore, Billerica, MA, USA) was performed in HFL1 cells. In brief, cells were lysed according to the guidebook of lysis buffer, and the lysates were subjected to incubation with magnetic beads conjugated with anti-Ago2 antibody (Abcam) or IgG antibody (Abcam). The immunoprecipitated RNAs were examined by qRT-PCR.
2.14. Statistical analysis
All assays were performed in triplicate. GraphPad Prism 7 was used for experimental statistics analysis. Significant differences were compared with Student’s t-tests or one-way analysis of variance (ANOVA). Data were expressed as means ± standard deviations (SD). p < 0.05 indicated statistical significance.
3. Results
3.1. Circ_0035796 depletion ameliorated TGF-β1-induced HFL1 cell proliferation, invasion, migration, inflammation and oxidative stress
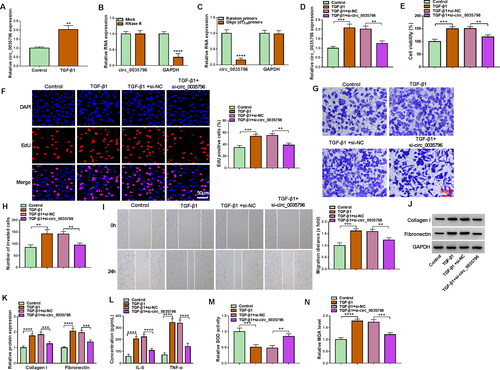
The results first showed that TGF-β1 treatment induced circ_0035796 expression in HFL1 cells (). We also identified the circular structure of circ_0035796 using RNase R, random primers and oligo(dT)18 primers. For instance, circ_0035796 was resistant to RNase R digestion, whereas linear GAPDH was sensitive to RNase R (). Random primers could amply both circ_0035796 and GAPDH, but oligo(dT)18 primers mainly amplified GAPDH (). Subsequently, we analysed the effects of circ_0035796 depletion on TGF-β1-induced HFL1 cell dysfunctions. The qRT-PCR data showed that TGF-β1 stimulation increased circ_0035796 expression, whereas the effect was relieved after transfection with siRNA of circ_0035796 (). As shown in , TGF-β1 treatment induced HFL1 cell viability and proliferation, while these effects were reversed when circ_0035796 expression was decreased. We also observed that TGF-β1 stimulation promoted HFL1 cell invasion and migration and the production of collagen I and fibronectin; however, these effects were restored by decreasing circ_0035796 expression (). Further, TGF-β1 stimulation increased IL-6, TNF-α and MDA production and decreased SOD activity, but these effects were remitted after circ_0035796 knockdown (). Thus, these data demonstrated that circ_0035796 depletion blocked TGF-β1-induced fibroblast activation.
Figure 1. Circ_0035796 depletion blocked TGF-β1-induced fibroblast activation. (A) The effect of TGF-β1 treatment on circ_0035796 expression was analysed by qRT-PCR in HFL1 cells. (B and C) The circular structure of circ_0035796 was identified using RNase R, random primers and oligo(dT)18 primers. HFL1 cells were divided into 4 groups, including the control group (untreated HFL1 cells), TGF-β1 group, TGF-β1 + si-NC group and TGF-β1 + si-circ_0035796 group, and circ_0035796 expression was analysed by qRT-PCR (D), cell viability by CCK-8 assay (E), cell proliferation by EdU assay (F), cell invasion by transwell invasion assay (G and H), cell migration by wound-healing assay (I), the protein expression of collagen I and fibronectin by Western blot (J and K), the production of IL-6 and TNF-α by ELISA (L), SOD activity by Superoxide dismutase activity assay kit (M) and MDA level by lipid peroxidation MDA assay kit (N). **p < 0.01, ***p < 0.001 and ****p < 0.0001.
3.2. Circ_0035796 acted as a miR-150-5p sponge in HFL1 cells
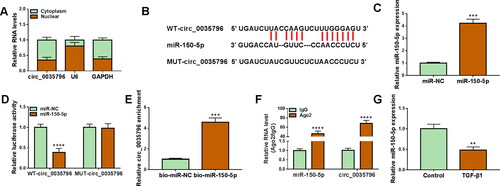
Subsequent data showed that circ_0035796 was mainly expressed in the cytoplasm of HFL1 cells, as presented in . Given that cytoplasmic circRNAs could act as miRNA sponges [Citation20], we searched for circ_0035796-associated miRNAs through Starbase online database. Given many miRNAs with circ_0035796-binding sites, we employed the miRNAs that were lowly expressed in pulmonary fibrosis patients and repressed pulmonary fibrosis development, including miR-506-5p, miR-942-5p and miR-150-5p. As shown in Figure S1A, circ_0035796 depletion significantly upregulated miR-942-5p and miR-150-5p expression, especially miR-150-5p expression. Thus, miR-150-5p was employed for the present work. As presented in , miR-150-5p, a candidate, contained circ_0035796-binding sites. Then, we performed a series of experiments to identify the association between the two. The success of miR-150-5p overexpression was shown in . Dual-luciferase reporter assay showed that miR-150-5p overexpression repressed the luciferase activity of wild-type reporter plasmid of circ_0035796 but not that of mutant reporter plasmid of circ_0035796 (). Moreover, circ_0035796 was dramatically enriched in the bio-miR-150-5p group compared with the bio-miR-NC group (). The results of RIP assay exhibited that both miR-150-5p and circ_0035796 were significantly higher in the Ago2 antibody group than in the IgG antibody group (). Further, we observed that miR-150-5p expression was dramatically downregulated in TGF-β1-treated HFL1 cells in comparison with the control group (). Thus, the above data suggested that circ_0035796 bound to miR-150-5p in HFL1 cells.
Figure 2. Circ_0035796 bound to miR-150-5p in HFL1 cells. (A) Circ_0035796 was mainly expressed in the cytoplasm of HFL1 cells. (B) The schematic illustration shows the binding sites of circ_0035796 for miR-150-5p. (C) The efficiency of miR-150-5p overexpression was analysed by qRT-PCR in HFL1 cells. (D-F) The association of circ_0035796 with miR-150-5p was identified by dual-luciferase reporter assay, RNA pull-down assay and RIP assay. (G) The effect of TGF-β1 treatment on miR-150-5p expression was determined by qRT-PCR in HFL1 cells. **p < 0.01, ***p < 0.001 and ****p < 0.0001.
3.3. MiR-150-5p depletion attenuated circ_0035796 knockdown-induced effects in TGF-β1-treated HFL1 cells
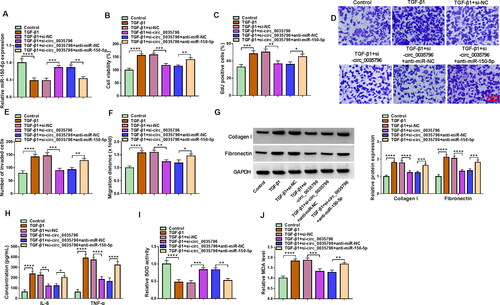
We then silenced both circ_0035796 and miR-150-5p in TGF-β1-induced HFL1 cells to determine whether the regulation of circ_0035796 on TGF-β1-induced fibroblast activation involved miR-150-5p. As shown in , circ_0035796 knockdown increased miR-150-5p expression in TGF-β1-treated HFL1 cells, whereas the effect was relieved after miR-150-5p expression was decreased. Subsequently, we observed that circ_0035796 knockdown repressed cell viability, cell proliferation, cell invasion and migration and decreased the expression of collagen I and fibronectin in TGF-β1-treated HFL1 cells; however, these effects were remitted by decreasing miR-150-5p expression (). In addition, the decreased production of IL-6, TNF-α and MDA and increased SOD activity by circ_0035796 silencing were relieved when miR-150-5p expression was downregulated in TGF-β1-treated HFL1 cells (). Taken together, all data demonstrated that circ_0035796 regulated TGF-β1-induced fibroblast activation through miR-150-5p.
Figure 3. Circ_0035796 regulated TGF-β1-induced fibroblast activation through miR-150-5p. HFL1 cells were divided into 6 groups, including control group, TGF-β1 group, TGF-β1 + si-NC group, TGF-β1 + si-circ_0035796 group, TGF-β1 + si-circ_0035796 + anti-miR-NC group or TGF-β1 + si-circ_0035796 + anti-miR-150-5p, and miR-150-5p expression was assessed by qRT-PCR (a), cell viability by CCK-8 assay (B), cell proliferation by EdU assay (C), cell invasion by transwell invasion assay (D and E), cell migration by wound-healing assay (F), the protein expression of collagen I and fibronectin by Western blot (G), the production of IL-6 and TNF-α by ELISA (H), SOD activity by Superoxide dismutase activity assay kit (I) and MDA level by lipid peroxidation MDA assay kit (J). *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
3.4. MiR-150-5p combined with L1CAM in HFL1 cells
Starbase database was employed to predict the target gene of miR-150-5p. Given many mRNAs with miR-150-5p-binding sites, the present study employed the mRNAs that were highly expressed in pulmonary fibrosis patients and promoted pulmonary fibrosis development, including COL1A1, ZEB1, STAT3, FOXM1, FOXK2, SP1, TGFBR1, and L1CAM. The results then revealed that miR-150-5p introduction decreased COL1A1, STAT3, SP1 and L1CAM expression, especially L1CAM expression (Figure S1B). Therefore, the present work selected L1CAM as a study subject. As shown in , L1CAM, a candidate, potentially bound to miR-150-5p. Subsequently, miR-150-5p introduction significantly repressed the luciferase activity of WT-L1CAM 3'UTR rather than that of MUT-L1CAM 3'UTR (). As analysed by RNA pull-down assay, bio-miR-150-5p dramatically enriched L1CAM compared with bio-miR-NC (). Moreover, we observed that both miR-150-5p and L1CAM were higher in the Ago2 antibody-induced silencing complexes than in the control group (). Further, L1CAM protein expression was significantly upregulated in the TGF-β1-treated HFL1 in comparison with the control group (). Thus, the above findings demonstrated that miR-150-5p combined with L1CAM in HFL1 cells.
Figure 4. MiR-150-5p combined with L1CAM in HFL1 cells. (A) the schematic illustration showed the complementary sites of miR-150-5p with L1CAM. (B-D) The association of miR-150-5p with L1CAM was identified by dual-luciferase reporter assay, RNA pull-down assay and RIP assay. (E) The effect of TGF-β1 treatment on L1CAM expression was determined by Western blot in HFL1 cells. ***p < 0.001 and ****p < 0.0001.
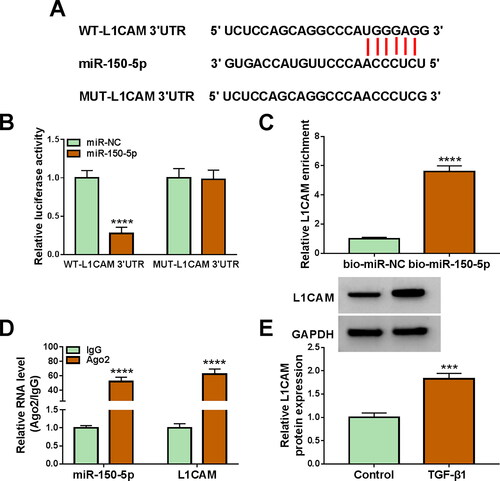
3.5. L1CAM overexpression relieved miR-150-5p-mediated effects in TGF-β1-treated HFL1 cells
The study continued to analyse whether miR-150-5p regulated TGF-β1-induced fibroblast activation by targeting L1CAM. To this end, we overexpressed miR-150-5p and L1CAM in TGF-β1-treated HFL1 cells and then analysed the subsequent effects on cell proliferation, invasion, migration, inflammation and oxidative stress. As presented in , miR-150-5p overexpression decreased L1CAM production in the cells, whereas these effects were attenuated after L1CAM introduction. Subsequently, miR-150-5p-induced inhibition in cell viability, proliferation, invasion, migration and the expression of collagen I and fibronectin was attenuated by increasing L1CAM expression in TGF-β1-stimulated HFL1 cells (). Consistently, ectopic L1CAM expression restored the inhibitory effects of miR-150-5p introduction on IL-6, TNF-α and MDA production and the promoting effect on SOD activity in TGF-β1-stimulated HFL1 cells (). Moreover, the data showed that L1CAM silencing attenuated TGF-β1 treatment-induced promoting effects on L1CAM expression, cell viability, cell proliferation, cell invasion, cell migration and collagen I, fibronectin, IL-6, and TNF-α production (Figure S2A-I). As presented in Figure S2J and K, TGF-β1 treatment inhibited SOD activity and promoted MDA production, whereas these effects were rescued after L1CAM depletion. Collectively, these data demonstrated that miR-150-5p combined with L1CAM to regulate TGF-β1-induced fibroblast activation.
Figure 5. MiR-150-5p combined with L1CAM to regulate TGF-β1-induced fibroblast activation. HFL1 cells were divided into 6 groups, control group, TGF-β1 group, TGF-β1 + miR-NC group, TGF-β1 + miR-150-5p group, TGF-β1 + miR-150-5p + pcDNA group and TGF-β1 + miR-150-5p + L1CAM group, and L1CAM expression was assessed by Western blot (A), cell viability by CCK-8 assay (B), cell proliferation by EdU assay (C), cell invasion by transwell invasion assay (D and E), cell migration by wound-healing assay (F), the protein expression of collagen I and fibronectin by Western blot (G), the production of IL-6 and TNF-α by ELISA (H), SOD activity by Superoxide dismutase activity Assay kit (I) and MDA level by lipid peroxidation MDA assay kit (J). *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
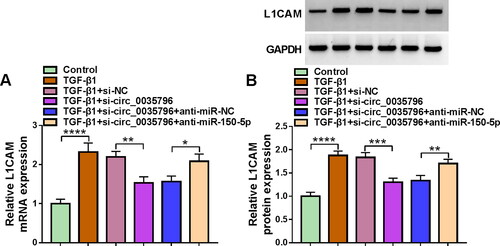
3.6. Circ_0035796 silencing repressed L1CAM production through miR-150-5p in TGF-β1-stimulated HFL1 cells
Based on the above results, we further analysed the associations among circ_0035796, miR-150-5p and L1CAM by transfecting siRNA of circ_0035796 and inhibitors of miR-150-5p into TGF-β1-stimulated HFL1 cells. As shown in , circ_0035796 knockdown repressed L1CAM mRNA and protein expression in the cells, whereas the effects were relieved after miR-150-5p knockdown, suggesting that circ_0035796 controlled L1CAM expression through miR-150-5p in TGF-β1-treated HFL1 cells.
Figure 6. Circ_0035796 controlled L1CAM expression through miR-150-5p in TGF-β1-treated HFL1 cells. HFL1 cells were divided into 6 groups, including control group, TGF-β1 group, TGF-β1 + si-NC group, TGF-β1 + si-circ_0035796 group, TGF-β1 + si-circ_0035796 + anti-miR-NC group or TGF-β1 + si-circ_0035796 + anti-miR-150-5p, and L1CAM expression was assessed by qRT-PCR (A) and Western blot (B). *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
4. Discussion
CircRNA is a single-stranded molecule that may provide approaches for disease therapy. In recent years, circRNA has been identified in fibrotic disorders [Citation21]. As reported, circ_0006916 silencing increased the levels of M1 molecules IL-1β and TNF-αand decreased M2 molecule TGF-β1 expression [Citation22], suggesting the contribution of circ_0006916 to silicosis and pulmonary fibrosis development. Cheng et al. indicated that circ_0058493 was a potential novel biomarker for silicosis and idiopathic pulmonary fibrosis [Citation23]. Although circRNA appears to serve important parts in pulmonary fibrosis [Citation11], its specific actions remain largely unknown. This study aimed to analyse circ_0035796 role in pulmonary fibrosis development and the detailed mechanism. The study showed that circ_0035796 depletion repressed TGF-β1-caused pulmonary fibrosis through the miR-150-5p/L1CAM pathway in vitro.
As determined in the study, circ_0035796 was upregulated in TGF-β1-exposed foetal lung fibroblast 1 cells and had a circular structure. After lung injury, fibroblasts are activated to proliferate and rebuild extracellular matrix to regulate wound healing for tissue repair [Citation24]. Moreover, at the site, fibroblast-to-myofibroblast transition can produce extracellular matrix components, which can promote wound healing [Citation25]. Based on these theories, we analysed TGF-β1-induced foetal lung fibroblast 1 cell proliferation and motility after transfection with si-circ_0035796. As a result, TGF-β1 induced cell proliferation, migration and invasion, which was relieved by decreasing circ_0035796 expression. Additionally, fibroblasts can lead to extracellular matrix accumulation (collagen I and collagen III) [Citation26]and can express high level of fibronectin. In the present work, TGF-β1 treatment increased collagen I and fibronectin expression in HFL1 cells, whereas these effects were attenuated after circ_0035796 knockdown. Further, we found that circ_0035796 silencing restored the promoting effects of TGF-β1 exposure on inflammation and oxidative stress. These findings suggested that circ_0035796 depletion blocked TGF-β1-induced fibroblast activation.
Cytoplasmic circRNA can serve as a miRNA sponge, further inhibiting miRNA-target interactions and protein function [Citation20]. Our data showed that circ_0035796 mainly functioned in the cytoplasm. Subsequently, we identified miR-150-5p as a target miRNA of circ_0035796. Considerable research has demonstrated the involvement of the miRNA in pulmonary diseases. For example, miR-150-5p was involved in the development of lung cancer [Citation27]. An early study reported that the decreased expression of miR-150-5p predicted poor survival rates of patients with chronic obstructive pulmonary disease [Citation17]. In particular, miR-150-5p was downregulated in transforming growth factor-β1-exposed HFL1 cells and combined with long noncoding RNA ZFAS1 and solute carrier family 38 member1 to repress HFL1 cell viability, cell migration, inflammatory cytokine secretion and MDA level [Citation19]. Consistently, we observed the downregulation of miR-150-5p in transforming growth factor-β1-exposed HFL1 cells and the inhibitory effects of miR-150-5p on cell viability, migration, inflammatory cytokine IL-6 and TNF-α secretion and MDA level in transforming growth factor-β1-exposed HFL1 cells. We also found miR-150-5p exhibited repressing roles in cell proliferation, cell invasion, SOD activity, and the expression of collagen I and fibronectin in TGF-β1-treated HFL1 cells. Further, circ_0035796/miR-150-5p axis regulated TGF-β1-induced fibroblast activation.
L1CAM is a transmembrane glycoprotein that is highly relevant for nervous system development and tumour tumorigenesis [Citation28]. Previous studies have demonstrated that the protein interacts with different binding partners in cis or trans [Citation29]. In addition, L1CAM can bind to cell adhesion molecules and integrins, and its cytoplasmic tail is capable of interacting with the cytoskeletal proteins ankyrin [Citation30,Citation31]. As reported, L1CAM contributed to lung cancer cell metastatic potential [Citation32]. Recently, Li et al. explained that L1CAM could increase epithelial-mesenchymal transition process in TGF-β1-induced A549 cells through the lncRNA cardiac hypertrophy related factor/miR-146a/L1CAM axis [Citation33]. In terms of mechanism, it has been demonstrated that L1CAM increases metastasis by inducing gelatinase expression [Citation34]. Our data confirmed miR-150-5p targeted L1CAM and that L1CAM expression was increased in TGF-β1-stimulated HFL1 cells. Moreover, ectopic L1CAM expression relieved miR-150-5p-mediated effects in TGF-β1-stimulated HFL1 cells, and L1CAM depletion attenuated TGF-β1-mediated effects in HFL1 cells. These data indicated that the miR-150-5p/L1CAM pathway regulated TGF-β1-induced pulmonary fibrosis in vivo.
However, the authors should validate the in vitro data using mouse model assays and determined which was the main cause of TGF-β1-induced fibroblast activation in further investigations, although it was complicated. In addition, other downstream mechanisms of circ_0035796-mediated pulmonary fibrosis processes deserved to be analysed in the future except for the miR-150-5p/L1CAM pathway.
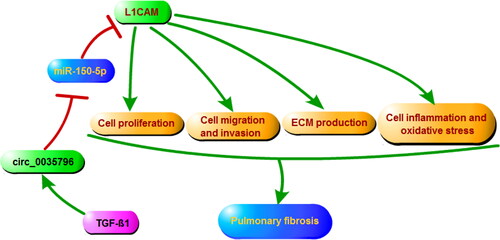
Taken together, circ_0035796 depletion ameliorated TGF-β1-induced fibroblast activation through the miR-150-5p/L1CAM pathway (), suggesting that the inhibitors of circ_0035796 might be a potential therapeutic option for pulmonary fibrosis.
Supplemental Material
Download MS Word (834.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Meyer KC. Pulmonary fibrosis, part I: epidemiology, pathogenesis, and diagnosis. Expert Rev Respir Med. 2017;11(5):1–10.
- Savin IA, Zenkova MA, Sen’kova AV. Pulmonary fibrosis as a result of acute lung inflammation: molecular mechanisms, relevant in vivo models, prognostic and therapeutic approaches. Int J Mol Sci. 2022;23:14959.
- Zhang J-X, Lu J, Xie H, et al. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019;10(3):182–182.
- Kinoshita T, Goto T. Molecular mechanisms of pulmonary fibrogenesis and its progression to lung cancer: a review. Int J Mol Sci. 2019;20:1461.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Chen Z, Song M, Wang T, et al. Role of circRNA in E3 modification under human disease. Biomolecules. 2022;12:1320.
- Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274.
- Li C, Wang Z, Zhang J, et al. Crosstalk of mRNA, miRNA, lncRNA, and circRNA and their regulatory pattern in pulmonary fibrosis. Mol Ther Nucleic Acids. 2019;18:204–218.
- Xu Q, Cheng D, Li G, et al. CircHIPK3 regulates pulmonary fibrosis by facilitating glycolysis in miR-30a-3p/FOXK2-dependent manner. Int J Biol Sci. 2021;17(9):2294–2307.
- Qi F, Li Y, Yang X, et al. Hsa_circ_0044226 knockdown attenuates progression of pulmonary fibrosis by inhibiting CDC27. Aging. 2020;12(14):14808–14818.
- Li R, Wang Y, Song X, et al. Potential regulatory role of circular RNA in idiopathic pulmonary fibrosis. Int J Mol Med. 2018;42(6):3256–3268.
- Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Disease Models & Mechanisms. 2021;14:dmm047662.
- Climent M, Viggiani G, Chen YW, et al. MicroRNA and ROS crosstalk in cardiac and pulmonary diseases. Int J Mol Sci. 2020;21:4370.
- Çakmak HA, Demir M. MicroRNA and cardiovascular diseases. Balkan Med J. 2020;37(2):60–71.
- Mo Y, Zhang Y, Wan R, et al. miR-21 mediates nickel nanoparticle-induced pulmonary injury and fibrosis. Nanotoxicology. 2020;14(9):1175–1197.
- Liu H, He Y, Jiang Z, et al. Prodigiosin alleviates pulmonary fibrosis through inhibiting miRNA-410 and TGF-β1/ADAMTS-1 signaling pathway. Cell Physiol Biochem. 2018;49(2):501–511.
- Keller A, Ludwig N, Fehlmann T, et al. Low miR-150-5p and miR-320b expression predicts reduced survival of COPD patients. Cells. 2019;8:1162.
- Suetsugu T, Koshizuka K, Seki N, et al. Downregulation of matrix metalloproteinase 14 by the antitumor miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous cell carcinoma cells. Int J Oncol. 2018;52:913–924.
- Yang Y, Tai W, Lu N, et al. lncRNA ZFAS1 promotes lung fibroblast-to-myofibroblast transition and ferroptosis via functioning as a ceRNA through miR-150-5p/SLC38A1 axis. Aging (Albany NY). 2020;12(10):9085–9102.
- van Zonneveld AJ, Kölling M, Bijkerk R, et al. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol. 2021;17(12):814–826.
- Fang S, Guo H, Cheng Y, et al. circHECTD1 promotes the silica-induced pulmonary endothelial-mesenchymal transition via HECTD1. Cell Death Dis. 2018;9(3):396.
- Wu Q, Jiao B, Zhang Q, et al. Identification of circRNA expression profiles and the potential role of hsa_circ_0006916 in silicosis and pulmonary fibrosis. Toxicology. 2023;483:153384.
- Cheng Z, Zhang Y, Wu S, et al. Peripheral blood circular RNA hsa_circ_0058493 as a potential novel biomarker for silicosis and idiopathic pulmonary fibrosis. Ecotoxicol Environ Saf. 2022;236:113451.
- Parimon T, Hohmann MS, Yao C. Cellular senescence: pathogenic mechanisms in lung fibrosis. Int J Mol Sci. 2021;22:6214.
- Kendall RT, Feghali-Bostwick CA. Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol. 2014;5:123–123.
- Nho RS, Ballinger MN, Rojas MM, et al. Biomechanical force and cellular stiffness in lung fibrosis. Am J Pathol. 2022;192(5):750–761.
- Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. 2017;16(1):118.
- Hua T, Liu S, Xin X, et al. Prognostic significance of L1 cell adhesion molecule in cancer patients: a systematic review and meta-analysis. Oncotarget. 2016;7(51):85196–85207.
- Kiefel H, Bondong S, Hazin J, et al. L1CAM: a major driver for tumor cell invasion and motility. Cell Adh Migr. 2012;6(4):374–384.
- Brümmendorf T, Kenwrick S, Rathjen FG. Neural cell recognition molecule L1: from cell biology to human hereditary brain malformations. Curr Opin Neurobiol. 1998;8(1):87–97.
- Herron LR, Hill M, Davey F, et al. The intracellular interactions of the L1 family of cell adhesion molecules. Biochem J. 2009;419(3):519–531.
- Liu X, Min S, Wu N, et al. miR-193a-3p inhibition of the slug activator PAK4 suppresses non-small cell lung cancer aggressiveness via the p53/slug/L1CAM pathway. Cancer Lett. 2019;447:56–65.
- Li J, Jiang ZZ, Li YY, et al. LncRNA CHRF promotes TGF-β1 induced EMT in alveolar epithelial cells by inhibiting miR-146a up-regulating L1CAM expression. Exp Lung Res. 2021;47(4):198–209.
- Hauser S, Bickel L, Weinspach D, et al. Full-length L1CAM and not its Δ2Δ27 splice variant promotes metastasis through induction of gelatinase expression. PLoS One. 2011;6(4):e18989.