Abstract
The human intestinal epithelium has an impressive ability to respond to insults and its homeostasis is maintained by well-regulated mechanisms under various pathophysiological conditions. Nonetheless, acute injury and inhibited regeneration of the intestinal epithelium occur commonly in critically ill surgical patients, leading to the translocation of luminal toxic substances and bacteria to the bloodstream. Effective therapies for the preservation of intestinal epithelial integrity and for the prevention of mucosal hemorrhage and gut barrier dysfunction are limited, primarily because of a poor understanding of the mechanisms underlying mucosal disruption. Noncoding RNAs (ncRNAs), which include microRNAs (miRNAs), long ncRNAs (lncRNAs), circular RNAs (circRNAs), and small vault RNAs (vtRNAs), modulate a wide array of biological functions and have been identified as orchestrators of intestinal epithelial homeostasis. Here, we feature the roles of many important ncRNAs in controlling intestinal mucosal growth, barrier function, and repair after injury—particularly in the context of postoperative recovery from bowel surgery. We review recent literature surrounding the relationships between lncRNAs, microRNAs, and RNA-binding proteins and how their interactions impact cell survival, proliferation, migration, and cell-to-cell interactions in the intestinal epithelium. With advancing knowledge of ncRNA biology and growing recognition of the importance of ncRNAs in maintaining the intestinal epithelial integrity, ncRNAs provide novel therapeutic targets for treatments to preserve the gut epithelium in individuals suffering from critical surgical disorders.
Introduction
Maintaining gut epithelial integrity is a complex process that demands expeditious alterations in the gene expression patterns of intestinal epithelial cells. This plasticity enables these cells to survive, proliferate, and migrate in times of pathophysiologic stress. These principles are particularly important in surgical patients whose intestines must recover and adapt following operative intervention as well as accompanying stressors such as dependence on total parenteral nutrition (TPN). After injury, inhibition of mucosal renewal in the intestine often occurs which can result in sepsis, multiple organ damage, and ultimately death. Despite this, supportive care remains the standard, and there are limited therapies that target restoration of intestinal epithelial regeneration and barrier function in patients with critical surgical diseases.
The intestinal epithelium quickly alters gene expression patterns to respond to stress by utilizing posttranscriptional regulation, including the alteration of mRNA stability and translation. Noncoding RNAs (ncRNAs) consist of multiple subtypes of transcripts, including microRNAs (miRNAs), small ncRNAs, long ncRNAs (lncRNAs), and circular RNAs (circRNAs). miRNAs bind directly to cis-elements found at 3′-untranslated regions (3′-UTRs) of their target mRNAs to either increase or decrease posttranscriptional gene expression [Citation1]. Evidence is accumulating that suggests the importance of these mechanisms in intestinal cell recovery and homeostasis following injury [Citation2–5]. In fact, defects in the pathways by which noncoding RNAs control mRNA stability and translation culminate in deficient epithelial renewal and barrier function leading to the sequalae of many critical surgical disorders [Citation6].
In this review article, we intend to outline the essential function of various ncRNAs in directing intestinal mucosal growth and adaptation in several pathophysiologic conditions found in surgical patients. We will also detail the mechanisms by which ncRNAs influence the stability and translation of target mRNAs through their interactions with each other and with RNA-binding proteins (RBPs).
Intestinal mucosal adaptation in response to surgical stress
The intestinal epithelium is one of three layers of the mucosa and is comprised of four major cells types—predominately absorptive enterocytes (80%), goblet cells, Paneth cells, and enteroendocrine cells (EECs) [Citation7,Citation8]. This intestinal lining is highly specialized for its dual purpose of absorbing nutrients while simultaneously blocking translocation of harmful bacteria into the bloodstream. Specialized EECs also secrete a large variety of gut hormones that regulate gastrointestinal (GI) activity and nutritional absorption [Citation9]. During weeks 9–10 of human gestation, columnar epithelium of the intestine invaginates to form villous structures and crypts of Lieberkuhn [Citation10]. Cellular differentiation along this crypt-villus axis is maintained by multiple signals including Wnt pathway [Citation11]. Constantly dividing intestinal stem cells (ISCs) located within the crypts generate bipotent progenitors which differentiate into absorptive or secretory offspring as they ascend vertically along the crypt-villus axis toward the intestinal lumen [Citation10,Citation11].
In response to pathological stress, intestinal epithelial cells (IECs) rapidly alter their gene expression [Citation12, Citation13], allowing undifferentiated IECs to replicate quickly within the proliferative zone of the crypts and differentiate as they migrate toward the intestinal lumen to take the place of lost or missing cells [Citation14]. To maintain constant epithelial renewal in stressful environments, the pluripotent LGR5+ stem cell can be replenished by neighboring lineage-restricted cells when LGR5+ cells are inactivated or depleted [Citation15]. Knowledge of the mechanisms of intestinal epithelial regeneration and differentiation is essential for understanding how the human intestinal mucosa adapts and recovers in patients with critical surgical diseases.
Following massive intestinal resection, the remaining small bowel undergoes “intestinal adaptation” to counteract lost surface area. Immediately after resection, crypt cells proliferate to increase crypt depth, villus height, tissue oxygenation, and blood flow [Citation10,Citation16]. The structural outcome following this process is hyperplasia, angiogenesis, bowel dilation, and bowel elongation. Studies from pediatric patients show that villus height and crypt depth increase by ∼32% and ∼22% at 74 days following resection, respectively [Citation17]. Paneth cell abundance within crypts also increases markedly 12 hours after resection and this change is sustained up to 28 days thereafter. The hypertrophy of the muscularis mucosa and propria also occurs quickly after resection, which results in augmented smooth muscle thickness, prominent mitochondria and sarcoplasmic reticulum, and a greater number of muscle cell nexuses [Citation10]. This adaptive process is strictly regulated by a myriad of factors such as IGF-1, PDGF, TNF-1, and FGF-10 [Citation10,Citation18,Citation19].
Functional changes after massive bowel resection include an increase in proteins involved in the transport and exchange of nutrients, electrolytes, and water, as well as a delay in transit time [Citation20]. Several signaling pathways, specifically epidermal growth factor (EGF) and B-cell lymphoma 2 (Bcl-2) receptors, are affected by bowel resection and thus govern this process [Citation21]. The adapting intestine is more metabolically active as shown by increased levels of ion channel activity, transporter proteins, transcriptional factors, DNA-binding protein receptors, RBPs, and cytoskeleton proteins [Citation22].
Massive small bowel resection also results in a significant change in the release of gut hormones. Glucagon-like peptide 2 (GLP-2) is produced by EECs of the intestinal epithelium and its expression levels depend on nutrient ingestion. After small bowel resection, GLP-2 levels increase by ∼30%, which promotes mucosal adaptation primarily by stimulating polyamine metabolism [Citation23]. When GLP-2 is administered exogenously, morphologic changes in the small bowel include increased length, mucosal thickness, villus height, and mucosal surface area [Citation24]. These changes in gut morphology are also apparent in other surgical states such as gut bypass surgery, after which GLP-2 levels increase resulting in restored absorptive mucosal surface [Citation25].
When more than half of the small intestine is removed, the resultant condition is known as short gut syndrome, which commonly requires chronic TPN. Because the presence of food is a strong pro-proliferative stimulus of the gut epithelium, chronic TPN inhibits mucosal growth in the intestine at least in part by decreasing FGF release [Citation26]. Furthermore, these surgical patients commonly experience states of polyamine depletion either due to loss of dietary polyamines during TPN use or from increased excretion postoperatively, the consequences of which will be detailed throughout this review [Citation12,Citation27]. In the upcoming sections, we will detail how many of these integral postoperative signaling pathways are regulated under the influence of ncRNAs.
MicroRNAs in gut mucosal growth and adaptation
miRNAs are small ncRNAs which influence translation and degradation of targeted mRNAs by interacting directly with their 3′-UTR [Citation28]. They are known to regulate more than 30% of mammalian gene transcripts [Citation29]. Emerging evidence indicates that miRNAs regulate intestinal mucosal regeneration and adaptation, and that control of miRNAs is essential for maintaining the intestinal epithelium homeostasis. miRNA levels are rapidly altered in surgical states such as bowel resection and bariatric surgery [Citation30,Citation31]. Our group has generated profiles of global miRNA expression in small intestine mucosa and cultured IECs which demonstrate high expression levels of multiple miRNAs, particularly miRNA-222, miRNA-29b, miRNA-322/503, and miR-195, the functions of which we will detail here. Levels of these miRNAs also vary dramatically in other pathological stress states such as polyamine depletion and food starvation. Here, we briefly outline the unique roles of miRNAs that are known to function in small intestine mucosal regeneration, barrier function, and adaptation in critical surgical conditions.
miR-222
miR-222, encoded on the X-chromosome, is conserved across species [Citation32]. In the intestinal epithelium, miR-222 is highly expressed and its levels change considerably in times of pathologic stress such as starvation or mucosal injury [Citation33,Citation34]. In cultured IECs, polyamine depletion increases cellular miR-222 and strengthens miR-222 interaction with the RBP CUG-binding protein 1 (CUGBP1) [Citation33]. Increased association of miR-222 with CUGBP1 diminishes the translation of cyclin-dependent kinase 4 (CDK4), leading to an inhibition of IEC proliferation. miR-222 overexpression also decreases levels of zipcode binding protein-1 (ZBP1) and phospholipase C-g1 (PLCg1) in IECs; two proteins intimately involved in gut mucosal regeneration and rapid epithelial restitution following an insult [Citation34].
By using intestine epithelial tissue-specific miR-222 transgenic (miR222-Tg) mice, we have further investigated the in vivo function of miR-222 in the intestinal epithelium. We found that miR222-Tg mice did not demonstrate alterations in body weight or gross GI morphology, but they do experience inhibited mucosal growth in the small bowel [Citation35]. Congruent with results observed in cultured IECs, the mucosal abundance of proliferative markers, including CDK4, Ki67, and PCNA, decreases in the small bowel of miR222-Tg mice relative to control littermates [Citation35]. Intestine specific miR-222 overexpression does not inherently induce mucosal damage or cellular demise, but it does increase the susceptibility of intestinal epithelial cells to pathologic stress. Ectopic expression of miR-222 also delays mucosal restoration following mesenteric ischemia/reperfusion (I/R)-induced injury and exacerbates intestinal barrier dysfunction secondary to cecal ligation and puncture (CLP) [Citation35].
The mRNA encoding Wnt receptor Frizzled-7 (FZD7) has been identified as a novel target of miR-222, and local increases in miR-222 expression in the intestinal epithelium downregulate the Wnt signaling pathway by repressing expression of FZD7 [Citation35]. This relationship accounts in part for the inhibited mucosal growth, nonhealed wounds, and intestinal barrier dysfunction accompanying miR-222 upregulation secondary to polyamine depletion in surgical patients, as well as in these models of surgical stress.
miR-29b
miR-29b is the most highly expressed constituent of the miR-29 family. miR-29b regulates cell growth and apoptosis and is implicated in various cellular tasks and certain malignancies through its modulation of target mRNAs and various cell signaling pathways [Citation36,Citation37]. miR-29b is essential in the adaptive immune response via interactions with interferon-γ and is implicated in the etiology of inflammatory bowel disease (IBD), wherein loss of its anti-fibrotic and anti-inflammatory properties leads to inflammation and stricture [Citation38,Citation39].
Our group has explored the involvement of miR-29b in intestinal epithelial homeostasis utilizing mouse models including food starvation and polyamine depletion. We have demonstrated increases in the levels of miR-29b during these times of stress, coinciding with inhibited mucosal growth and dysfunction of the intestinal barrier [Citation40]. Conversely, miR-29b silencing in mice stimulates regeneration of the intestinal epithelium and promotes gut barrier integrity. miR-29b exhibits this effect by targeting and repressing the mRNA encoding CDK2, ultimately resulting in G1 phase growth arrest of IECs [Citation40]. In intestinal epithelial tissues, miR-29b abundance is tightly controlled by JunD at the transcriptional level [Citation41,Citation42]. JunD is an activator protein (AP)-1 transcription factor and interacts directly with the miR-29b1 promoter to stimulate its expression. JunD inhibits mucosal growth and weakens gut barrier integrity by elevating levels of p21 and by decreasing CDK4 and tight junction proteins [Citation41]. miR-29b also influences mucosal regeneration through repression of the Wnt co-receptor LRP6 and the RBP HuR, thus inhibiting IEC proliferation and impairing gut barrier integrity [Citation40,Citation43]. miR-29b also targets Men1 mRNA to repress its expression posttranscriptionally, thereby lowering Menin protein and altering susceptibility of IECs to apoptosis [Citation44]. Given these findings, miR-29b represents an interesting target for pharmacotherapy to improve gut mucosal regeneration and barrier function.
miR-195
miR-195 is conserved throughout evolution, and it is abundant in the epithelial tissue of the intestine [Citation45]. miR-195 regulates cell proliferation through control of the expression of various molecules. Its levels are downregulated in numerous human cancers, and upregulated in states of mucosal atrophy secondary to starvation [Citation45–48]. Within intestinal epithelial tissues, miR-195 functions cooperatively with CUGBP1 to repress the translation of insulin-like growth factor 2 receptor (IGF2R), thereby impairing mucosal regeneration and adaptation [Citation49].
miR-195 also influences intestinal wound healing after surgical insult through exerting control over rapid epithelial restitution, the process by which epithelial cells migrate over the wounded area and re-differentiate to close the defect [Citation45]. miR-195 interacts stim1 mRNA to destabilize and prevent its translation, which prevents epithelial restitution after wounding by reducing Ca2+ influx. miR-195 association with stim1 is prevented by HuR, and the miR-195-mediated inhibition of epithelial restitution is consistently blocked by HuR overexpression [Citation45]. Further investigation reveals that miR-195 and HuR compete to bind to the stim1 3′-UTR and that the interaction between miR-195 and HuR controls stability of the stim1 mRNA [Citation45].
Another mechanism by which miR-195 decreases epithelial restitution is through the regulation of actin-related proteins (ARPs). miR-195 overexpression destabilizes the ARP-2 mRNA, decreases cellular ARP-2 levels, and slows down IEC migration over wounded regions through inhibiting formation of F-actin stress fibers [Citation50]. Moreover, miR195-Tg mice also exhibit an increase in vulnerability of the intestinal barrier to lipopolysaccharide (LPS).
miR-195 levels in the intestinal epithelium are highly controlled by cellular polyamines and lncRNA uc.173 [Citation50,Citation51]. Polyamine depletion increases expression of miR-195 in IECs, whereas elevated levels of polyamines decrease miR-195 abundance. In addition, uc.173 associates directly with the primary miR-195 (pri-miR-195) to induce pri-miR-195 degradation [Citation51]. Together, these results imply that miR-195 negatively regulates homeostasis of the intestinal epithelium, and deregulated miR-195 in patients with critical surgical disorders disrupts renewal of the intestinal mucosa to cause gut barrier dysfunction.
miR-125a/214
miR-125a and miR-214 levels increase notably (∼2.4-fold and ∼3.2-fold, respectively), following massive intestine resection in rats [Citation29]. Further experiments demonstrate that the increased levels of miR-125a and 214 contribute to the increased apoptosis seen following small bowel resection and that increasing levels of miR-125a reduces expression of the antiapoptotic gene Mcl1 of the Bcl-2 family [Citation52]. These studies align with previous works that show the involvement of EGF signaling and other Bcl-2 family members in regulating apoptosis during intestinal adaptation [Citation29,Citation53]. As mentioned earlier, GLP-2 has intestinotrophic effects and its levels increase following massive bowel resection. Treatment with GLP-2 increases regeneration of the intestinal mucosa in short bowel rats by downregulating the content of exosomal miR-125a/b [Citation54]. This exemplifies how known therapies for surgical diseases such as short gut syndrome can be further explained using knowledge of miRNA pathways.
miR-503/322
miR-503 is highly conserved in a wide variety of species and it is clustered with miR-322 on the X chromosome [Citation55,Citation56]. Both miR-503 and miR-322 are involved in various human pathologies including cardiovascular disease and cancer [Citation57]. In the intestinal epithelium, miR-503/322 expression decreases markedly in the state of mucosal atrophy following food starvation and polyamine depletion [Citation1,Citation58]. Increasing the levels of miR-503 represses CUGBP1 expression and induces vulnerability of IEC cells to apoptosis by increasing c-Myc [Citation58]. Mechanistically, miR-503 recruits the Cugbp1 mRNA to P-bodies where non-translating mRNAs are harbored and targeted for deadenylation and degradation [Citation58]. miR-503 and miR-322 also regulate apoptosis by altering the expression of Smurf2, an ubiquitin ligase which targets certain proteins for degradation. Lowering miR-322/503 levels increase Smurf2 levels, ultimately increasing resistance of IECs to apoptosis [Citation56].
In summary, levels of tissue miRNAs in the gut mucosa are markedly affected by critical surgical stress, food starvation, and polyamine levels [Citation49–51]. miRNAs can directly bind to target mRNAs to function as mRNA destabilizers and/or repressors of mRNA translation, thus decreasing expression levels of various target genes (). RBPs such as HuR and CUGBP1 interact with miRNAs such as miR-195 and miR-222 and jointly regulate production of target mRNAs synergistically or antagonistically [Citation45,Citation49]. This evidence shows that altering levels of different miRNAs in patients with critical surgical disorders disrupts intestinal epithelium homeostasis and adaptation by impairing IEC survival, proliferation, migration, and cell-to-cell interaction. These pathways should be investigated for pharmacologic therapies. Although a comprehensive explanation of all known miRNA is beyond the scope of this paper, we have summarized other known miRNAs, including miR-16, miR-146a, miR-let7e-5p, miR-429, miR-21, miR-29a, miR-125b, miR-200c, and miR-144 [Citation59–66], which are relevant to gut mucosal function in .
Figure 1. miRNAs regulate the intestinal epithelium homeostasis by altering IEC survival, proliferation, migration, and cell-to-cell interaction. In this model, the levels of mucosal miRNAs are increased in the intestine after exposure to various pathological stresses. Increased miRNAs inhibit expression of their target genes by decreasing stability and translation of the mRNAs, thus leading to disruption of the gut epithelium homeostasis. RNA binding proteins (RBPs) such as HuR and CUGBP1 can interact with miRNAs including miR-195 and miR-222 and modulate their regulatory functions antagonistically or synergistically.
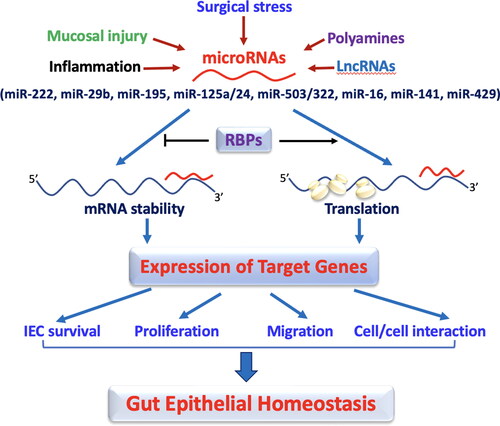
Table 1. Functions and targets of miRNAs in the intestinal epithelium.
LncRNAs in gut mucosal growth and adaptation
LncRNA are a subgroup of ncRNAs defined by length greater than 200 nucleotides. They participate in diverse cellular mechanisms and pathobiologies [Citation67–70]. LncRNA are transcribed by RNA polymerase II and share structural similarities with mRNAs. However, lncRNAs are unique from other regulatory and structural RNAs in that they are not well conserved across species, and their expression is cell type specific in response to different stresses [Citation2,Citation71]. LncRNAs function in various ways including chromatin reprogramming, cis-regulation at enhancers, and posttranscriptional regulation of mRNA processing [Citation68]. LncRNAs also act as molecular signals, decoys, RNA guides, and molecular scaffolds. A few functional lncRNAs have been identified and well investigated in humans thus far, though an increasing body of evidence indicates that lncRNAs modulate regeneration and adaptation of the intestinal epithelium. The dysregulation and mutations of lncRNAs are closely implicated in the pathogenesis of many gut mucosal disorders. Here, we outline several well described lncRNAs that have been evaluated and shown to play an essential role in maintaining homeostasis of the intestinal epithelium.
uc.173
A number of lncRNAs are transcribed from genomic ultraconserved regions (T-UCRs) which are absolutely conserved between orthologous regions of human, rat, and mouse genomes [Citation2]. uc.173 is a member of the T-UCR family and highly expressed in the intestinal epithelium [Citation71]. Expression levels of uc.173 decrease dramatically in mouse intestinal mucosa after exposure to food starvation, which is accompanied by inhibited intestinal epithelial renewal [Citation72]. Inhibition of uc.173 by treatment with its antagonist inhibits small bowel mucosal growth in mice. Interestingly, intestinal mucosal tissue samples collected from patients with Crohn’s disease demonstrate a notable decrease in uc.173 levels in comparison to control individuals.
In cultured IEC-6 cells, raising the uc.173 levels increases cell proliferation, while attenuating uc.173 prevents IEC division. Similarly, ectopically expressed uc.173 stimulates growth of primarily cultured intestinal organoids. uc.173 promotes growth of small bowel mucosa in part by decreasing miR-195, which we have previously described as a biologic disruptor of intestinal epithelial renewal [Citation49,Citation50]. Increasing endogenous miR-195 by uc.173 silencing with locked nucleic acid anti-uc.173 inhibits IEC proliferation. Mechanistically, uc.173 associates directly with pri-miR-195, resulting in an increase in miR-195 degradation [Citation72].
uc.173 also improves the integrity of the epithelial barrier by promoting production of the tight junction (TJ) protein claudin-1 [Citation42]. Decreasing the levels of cellular uc.173 by its silencer impairs gut barrier function in an in vitro model by specifically reducing the amount of claudin-1. In mice, decreased uc.173 expression results in high susceptibility of the intestinal barrier to septic stress induced by CLP. Exposure to CLP consistently lowers production of TJ proteins claudin-1 and claudin-3 in the intestinal mucosa, but these decreases in claudin-1 are more pronounced with anti-uc.173-treated mice. uc.173 increases translation of claudin-1 through interacting with miR-29b rather than directly associating with the claudin-1 mRNA. uc.173 functions as a decoy RNA for miR-29b which directly binds to claudin-1 mRNA to repress claudin-1 translation. Ectopic overexpression of uc.173 prevents the interaction of miR-29b with claudin-1 mRNA and rescues claudin-1 production in cells with high levels of miR-29b, thus restoring epithelial barrier function [Citation42]. Therefore, delivery of uc.173 is protective of the epithelial barrier.
uc.230
uc.230 is another member of the T-UCR family of lncRNAs and has been recognized as a major regulator of epithelial regeneration, apoptosis, and barrier integrity in the intestine [Citation2,Citation73]. The levels of uc.230 are increased in samples from individuals with ulcerative colitis, in mouse models of colitis and fasting, and in states of polyamine depletion [Citation2]. Decreasing cellular uc.230 prevents proliferation of IECs in vitro and inhibits growth of intestinal organoids ex vivo, thus leading to epithelial barrier dysfunction. Silencing uc.230 causes dysfunction of the epithelial barrier and increases vulnerability to apoptosis, while its overexpression proves to be protective. Attenuating uc.230 levels in mice enhances mucosal inflammatory injury and delays recovery of damaged mucosa in colon after administration of dextran sulfate sodium (DSS) in drinking water [Citation2]. Mechanistically, uc.230 increases CUGBP1 by functioning as a natural decoy RNA for miR-503, which associates with Cugbp1 mRNA to repress its translation. Together, uc.230 appears to protect the mucosa from further injury during times of inflammation by promoting epithelial renewal and decreasing susceptibility to apoptosis. These findings suggest that the uc.230/CUGBP1 axis is another potential therapeutic target for patients with gut mucosal inflammatory disorders.
SPRY4-IT1
LncRNA sprouty receptor kinase signaling antagonist 4-intronic transcript 1 (SPRY4-IT1) is derived from an intron of the SPRY4 gene and expressed in a variety of human cell types including the intestinal epithelium [Citation74,Citation75]. SPRY4-IT1 is primarily localized in cell cytoplasm, and its expression increases markedly in many human malignancies such as bladder, renal, gallbladder, and gastric cancer [Citation75,Citation76]. Our team has examined the involvement of SPRY4-IT1 in maintaining homeostasis of the intestinal epithelium. In samples of bowel tissue from patients with leaky gut, there are lower levels of SPRY4-IT1, as well as low levels of TJ proteins including claudin-1, claudin-3, occludin, and JAM-1 [Citation77]. Similarly, when SPRY4-IT1 is silenced in vitro, epithelial cells exhibit inhibited expression of the aforementioned proteins. Decreasing the levels of cellular SPRY4-IT1 also interrupts gut barrier function in an in vitro model, as demonstrated by a decrease in the values of transepithelial electrical resistance and an increase in paracellular permeability. In contrast, SPRY4-IT1 overexpression in gut mucosa protects the integrity of the intestinal barrier in septic mice induced by CLP. Mice exposed to CLP also exhibit lower levels of tissue claudin-1, claudin-3, occludin, and JAM-1 in the intestinal mucosa, but this effect is attenuated or counteracted completely by elevating mucosal SPRY4-IT1 levels.
Mechanistically, SPRY4-IT1 regulates production of TJ proteins posttranscriptionally by stabilizing and stimulating the translation of mRNAs encoding these TJ proteins via improvement of the association between the mRNAs and highly activated polyribosomes. Interestingly, the interactions between SPRY4-IT1 and TJ mRNAs are enhanced by the RBP HuR [Citation77]. This link makes SPRY4-IT1 a novel target for molecular therapies for critical surgical patients, such as sepsis, trauma, and thermal injury, who experience associated altered gut permeability.
H19
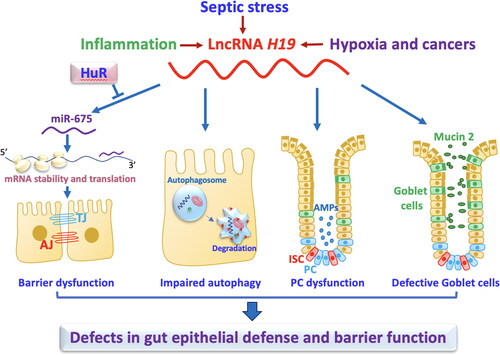
H19 is a 2.3-kb lncRNA transcribed from conserved imprinted H19/igf2 gene cluster located on chromosome 11p15.5 in humans [Citation78]. H19 levels are increased in murine colitis models and mucosal tissue samples from individuals with ulcerative colitis and sepsis [Citation79,Citation80]. In an in vitro system, overexpression of H19 impedes intestinal barrier function by inhibiting the production of TJ protein ZO-1 and AJ protein E-cadherin [Citation81]. H19 represses expression of ZO-1 and E-cadherin by reducing stability and translation of the ZO-1 and E-cadherin mRNAs.
Further study shows that increased H19 exhibits its effect on the barrier by increasing production of miR-675-3p and miR-675-5p [Citation81], two conserved miRNAs derived from H19 exon 1. This effect is counteracted by HuR, since HuR interacts with H19 and decreases miR-675 levels by preventing its processing from H19. Consistently, increasing HuR levels restores expression of ZO-1 and E-cadherin, thus protecting the epithelial barrier function in the epithelium overexpressing H19.
In addition to its effects on TJ and AJ proteins, it has been also reported that elevated H19 contributes to the pathogenesis of gut barrier dysfunction through debilitating function of Paneth cells and goblet cells and suppressing autophagy [Citation80]. In H19-knockout mice, both Paneth cells and goblet cells are preserved against septic stress. Furthermore, autophagy activity and intestinal barrier function are protected after exposure to CLP in H19-deficient mice. Intestinal organoids derived from H19-knockout mice have a greater number of Paneth cells and goblet cells, and they also are more tolerant to LPS than their control counterparts. In contrast, H19 upregulation in cultured epithelial cells prohibits rapamycin-induced autophagy and counteracts the rapamycin-induced protection of the epithelial barrier against LPS [Citation80].
Increased H19 in inflamed intestinal tissues is also shown to antagonize p53 function and inhibits activities of miR-34b and let-7 by altering the IL-22/STAT3 and cAMP-independent protein kinase-A pathways, thus contributing to the control of intestinal epithelial repair after injury [Citation79]. Although H19 functions as a let-7 sponge in other studies, reducing the cellular levels of let-7 does not independently lower the expression of ZO-1 and E-cadherin [Citation81].
Consistently, intestinal mucosal tissues from patients with adhesive small bowel obstruction also exhibit increased H19 expression that is positively correlated to aquaporin-3 (AQP3) and inversely proportional to miR-874. H19 acts as a competitor for miR-874 and leads to the stimulation of AQP3 expression [Citation82]. Taken together, these studies indicate that induced H19 contributes gut barrier dysfunction () and that attenuating the effect of H19 may prove protective against bacterial translocation in individuals with sepsis.
Figure 2. LncRNA H19 disrupts the intestinal defense and barrier function via distinct mechamisms. Induced levels of mucosal H19 in the intestine by septic stress and mucosal inflammation repress expression of tight junctions (TJs) and adherens junctions (AJs), impair autophagy, and disrupt function of Paneth cells (PCs) and goblet cells. H19 inhibits expression of ZO-1 and E-cadherin posttranscriptionally through release of miR-675 embedded in H19 exon 1, and this inhibition is prevented by HuR.
Gata6
LncRNA Gata6 serves a unique purpose in the regulation of gut adaptation and recovery through its influence on ISCs. Unlike uc.173 and uc.230 which are constitutively expressed in all IECs, transcriptome analysis conducted in sorted Lgr5+ and Lgr5– crypt cells isolated from the intestinal mucosa reveals that Gata6 levels are significantly elevated in adult intestinal epithelial cells when compared to other cell types [Citation83]. When the Gata6 locus is deleted from the mouse genome, ISCs lose their ability to differentiate; therefore, the mucosa is unable to regenerate following acute injury secondary to radiation [Citation83]. Mechanistically, Gata6 enhances ISC proliferation by activating WNT signals via stimulation of Ehf transcription and by recruiting the NURF complex onto the Ehf promoter. Gata6 expression is elevated in human colon cancers, and levels correlate positively with disease severity and prognosis in patients, thus making it a potential target for cancer therapy [Citation83]. It may conversely be a valuable target to encourage mucosal regeneration. This concept highlights the essential balance between epithelial recovery and the consequences of over-proliferation in the intestinal epithelium.
In summary, we have demonstrated how lncRNAs work alongside miRNA and RBPs in complex regulatory networks that control intestinal epithelial regeneration and adaptation in various pathological conditions. These molecules are an exciting potential therapeutic target for surgical recovery, inflammatory disorders, and oncologic therapies. In addition, several other lncRNAs, including BMP1, PlncRNA1, CCAT1, BC012900, and NAIL [Citation4,Citation84–87], listed in have been identified and investigated in the intestinal epithelium, but their roles and involvement in the process of recovery from critical surgical stress are not yet fully understood.
Table 2. Function of lncRNA in the intestinal epithelium.
CircRNAs in intestinal mucosal healing
CircRNAs are a varied class of endogenous ncRNAs that frequently have altered expression based in different tissue types, disease states, and developmental stages [Citation88]. Their covalently closed looped structures do not have 5′ to 3′ ends, and they are generated by an alternative splicing process known as back-splicing. The circular structure of circRNAs makes them uniquely resistant to degradation, resulting in a prolonged half-life when compared to linear RNAs. CircRNAs are commonly concentrated in cellular cytoplasm and participate in various biological mechanisms via functioning as miRNA sponges, protein scaffolds, decoys, and recruiters [Citation89,Citation90]. Each circRNA can have one or many miRNA-binding sites, and some are shown to harbor binding sites for numerous different miRNAs. CircRNAs also act in tandem with RBPs to regulate gene expression and are involved in many aspects of multiple cellular functions and pathologies in the intestinal epithelium [Citation89–91].
circHIPK3
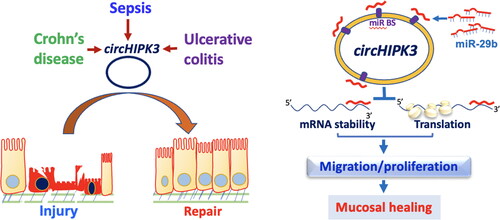
circHIPK3 originates from exon 2 of the HIPK3 gene via direct back-splicing and has been identified in many mammalian tissues including the intestine [Citation92,Citation93]. circHIPK3 controls cellular mechanisms in a cell-type-dependent manner primarily through its association with various miRNAs [Citation94]. Utilizing genome-wide profiling analysis, our group has identified ∼300 circRNAs, including circHIPK3, whose levels vary significantly in the gut mucosa of CLP-mice relative to sham mice [Citation93]. In mucosal samples from patients with sepsis or IBD, circHIPK3 levels decrease remarkably. In an in vitro model and in mice with intestinal mucosal injury, increasing the levels of cellular circHIPK3 improves intestinal epithelial repair after wounding, while circHIPK3 downregulation delays recovery of damaged mucosa. Inhibition of circHIPK3 also decreases IEC proliferation and growth of intestinal organoids. Mechanistically, circHIPK3 stimulates mucosal healing and promotes constant epithelial renewal in the intestine by altering miR-29b activity. circHIPK3 directly binds to miR-29b to decrease the availability of miR-29b, thus resulting in an increase in the levels of Rac1, Cdc42, and cyclin B1 ().
Figure 3. Model proposed to explain the influence of circHIPK3 upon intestinal epithelial repair after wounding. The levels of circHIPK3 are markedly altered in the intestinal mucosal tissues from patients with sepsis, Crohn’s disease, and ulcerative colitis. Increased circHIPK3 plays an essential role in intestinal mucosal repair after injury via interaction with miR-29b.
circPABPN1
CircPABPN1 is derived from the PABPN1 gene and shown to modulate PABPN1 levels through disrupting the interaction between HuR and PABPN1 mRNA [Citation95]. Our recent studies show that circPABPN1 regulates the intestinal epithelial-host defense and barrier integrity through interaction with HuR to influence autophagy [Citation96]. Overexpression of circPABPN1 stops the association of HuR with the Atg16l1 mRNA which culminates in reduction of the ATG16L1 protein and therefore prevents autophagy activation. This inhibition is specific, since circPABPN1 does not impact expression levels of other autophagy proteins such as ATG5. Interestingly, elevation of HuR levels recovers ATG16L1 expression in cells that overexpress circPABPN1, whereas decreasing the levels of cellular HuR and ectopically overexpressed circPABPN1 repress ATG16L1 expression synergistically. Human intestinal mucosal samples retrieved from patients with IBD display an elevated amount of circPABPN1 and lower HuR level, which is associated with a reduction in ATG16L1 protein and gut barrier dysfunction [Citation96]. Together these results suggest that circPABPN1 influences autophagy activation by altering ATG16L1 expression by affecting HuR function in the intestinal epithelium, ultimately altering intestinal epithelium-host defense and barrier function.
Cdr1as
Cdr1as is ubiquitous in the human body and shown to act as a sponge and antagonist for miR-7 [Citation91]. We have recently identified Cdr1as as a repressor of intestinal epithelial regeneration and defense [Citation97]. Cdr1as levels increase in mouse intestinal mucosa after colitis and septic stress, as well as in human intestinal mucosal samples from individuals with sepsis and IBD. Ablation of the Cdr1as locus from the mouse genome enhances renewal of the intestinal mucosa, promotes injury-induced epithelial regeneration, and protects the mucosa against DSS-induced colitis. More than 40 miRNAs, including miR-195, are differentially expressed among the intestinal mucosa of Cdr1as knockout vs littermate mice. Elevating the amount of Cdr1as inhibits intestinal epithelial recovery following wounding in cultured IECs and represses growth of intestinal organoids, but this inhibition is counteracted by miR-195 knockdown [Citation97].These exciting findings suggest that Cdr1as inhibits recovery of damaged mucosa in the intestine partially via its interaction with miR-195.
Small noncoding vault RNAs in gut barrier function
Vault RNAs (vtRNAs) are small ncRNAs with ∼100 nucleotides transcribed by RNA polymerase III [Citation98]. vtRNAs are highly expressed in many eukaryotes and they interact with giant cytoplasmic ribonucleoprotein particles termed vaults that contain major vault proteins (MVPs). vtRNAs also function independently of MVPs and are associated with many cellular processes, including mRNA splicing, nuclear transport, drug resistance, synaptogenesis, lysosome function, apoptosis, and tumorigenesis [Citation98,Citation99]. Humans express four vtRNA paralogs, including vtRNA1-1, vtRNA1-2, vtRNA1-3, and vtRNA2-1, while mice only produce one vtRNA. The levels of vtRNAs increase in response to stress, and a rise in free vtRNA levels modulates many cellular processes and functions. Recent studies reveal an important role of these abundant small vtRNAs in intestinal epithelium homeostasis and pathologies [Citation99].
vtRNA2-1 is a 108-nucleotide long ncRNA and can be incorporated into vault particles or remain free in the cytoplasm. We have recently reported that vtRNA2-1 functions as a novel orchestrator of the intestinal barrier via interaction with HuR [Citation3]. Samples of the intestine taken from individuals with IBD and from mice with colitis or sepsis express increased levels of vtRNAs relative to controls. Ectopically expressed vtRNA2-1 lowers the levels of TJ proteins claudin 1, occludin, and AJ E-cadherin and causes intestinal epithelial barrier dysfunction, whereas vtRNA2-1 silencing promotes gut barrier function. Increased vtRNA2-1 also decreases these junctional proteins in intestinal organoids, inhibits epithelial renewal, and causes Paneth cell defects ex vivo. Elevating the levels of tissue vtRNA2-1 in the intestinal mucosa elevates the susceptibility of the epithelial barrier to septic stress secondary to CLP in mice. Mechanistically, vtRNA2-1 interacts with HuR to hinder HuR binding to the mRNAs encoding claudin-1 and occludin proteins, thus decreasing their translation [Citation3]. This study suggests that vtRNA2-1 inhibits gut barrier function via suppressing HuR-facilitated translation of claudin-1 and occludin.
In addition, vtRNA1-1 is abundant in the intestinal epithelium and also implicated in the control of proliferation and autophagy in several lines of cancer cells [Citation100–102], but its exact role in preserving intestinal epithelium homeostasis and barrier function in stressful environments remains unknown. Vault RNAs are a relatively new field of study, and further details on their roles in the intestinal epithelium will surely emerge in the upcoming years.
Future directions and perspectives
We have outlined the numerous intricate ways by which ncRNAs exhibit posttranscriptional control of intestinal epithelial regeneration and adaptation, particularly the states of critical stress secondary to surgical disease. This work is one of the first of its kind to synthesize our knowledge of ncRNAs in the context of surgical pathology. These pathways are prime targets for the development of new clinical applications to treat patients with critical surgical disorders. Most ncRNAs, such as circRNAs and vtRNAs, demand further exploration, given their apparent roles in intestinal pathophysiology. There is a paucity of experiments utilizing human intestinal tissue from different surgical illnesses, and filling this gap is critical to fully understand the consequence of altered ncRNA expression on human disease with the ultimate goal of devising novel therapeutic strategies. Furthermore, the downstream consequences of mutations on ncRNA functional motifs and their regulatory domains on their capacity to influence other ncRNAs and/or RBPs and ultimately epithelial homeostasis must also be comprehensively investigated.
An appreciation of the exact mechanisms by which ncRNAs exert their influence on intestinal regeneration and adaptation is integral to understanding and developing novel therapies for the treatment for patients with various critical gut diseases [Citation102–106]. A well-known example of this is the application of GLP-2 to promote intestinal rehabilitation in patients with short gut syndrome via downregulation of miR-125a/b [Citation103]. This concept whereby ncRNAs are manipulated for the therapeutic benefit of patients with critical surgical diseases can be also generalized to applications in many other diseases. Although the development of ncRNA-guided therapeutics still faces many challenges, particularly in their specificity, deliverability, and tolerability [Citation104], there is not any doubt, with rapidly advancing knowledge of ncRNA biology and interactions between ncRNAs and RBPs, ncRNAs are exciting targets to safeguard intestinal epithelial integrity in individuals experiencing critical surgical disorders.
Acknowledgment
JY Wang is a Senior Research Career Scientist, Biomedical Laboratory Research & Development Service, US Department of Veterans Affairs.
| Abbreviations | ||
| IECs | = | intestinal epithelial cells |
| ncRNAs | = | noncoding RNAs |
| lncRNAs | = | long ncRNAs |
| RBPs | = | RNA-binding proteins |
| IBD | = | inflammatory bowel diseases |
| RRM | = | RNA recognition motif |
| miRNA | = | microRNA |
| CLP | = | cecal ligation and puncture |
| CRs | = | coding regions |
| UTRs | = | untranslated regions |
| circRNAs | = | circular RNAs |
| vtRNA | = | vaultRNA. |
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Wang J-Y, Xiao L, Wang J-Y. Posttranscriptional regulation of intestinal epithelial integrity by noncoding RNAs. Wiley Interdiscip Rev RNA. 2017;8(2):1. doi:10.1002/wrna.1399.
- Yu T-X, Kalakonda S, Liu X, et al. Long noncoding RNA uc.230/CUG-binding protein 1 axis sustains intestinal epithelial homeostasis and response to tissue injury. JCI Insight. 2022;7(19):e156612. doi:10.1172/jci.insight.156612.
- Ma X, Xiao L, Wen S, et al. Small noncoding vault RNA2-1 disrupts gut epithelial barrier function via interaction with HuR. EMBO Rep. 2022;24(2):e54925. doi:10.15252/embr.202254925.
- Zhuang M, Deng Y, Zhang W, et al. LncRNA Bmp1 promotes the healing of intestinal mucosal lesions via the miR-128-3p/PHF6/PI3K/AKT pathway. Cell Death Dis. 2021;12(6):595. doi:10.1038/s41419-021-03879-2.
- Xiao L, Gorospe M, Wang J-Y. Long noncoding RNAs in intestinal epithelium homeostasis. Am J Physiol Cell Physiol. 2019;317(1):C93–12. doi:10.1152/ajpcell.00092.2019.
- Chung HK, Rao JN, Wang J-Y.5.11. Regulation of gut barrier function by RNA-binding proteins and noncoding RNAs. In: Kenakin T, ed. Comprehensive Pharmacology. Elsevier; 2022:194–213. doi:10.1016/b978-0-12-820472-6.00059-1.
- Le Beyec J, Billiauws L, Bado A, et al. Short bowel syndrome: a paradigm for intestinal adaptation to nutrition? Annu Rev Nutr. 2020;40(1):299–321. doi:10.1146/annurev-nutr-011720-122203.
- Takahashi T, Fujishima K, Kengaku M. Modeling intestinal stem cell function with organoids. Int J Mol Sci. 2021;22(20):10912. doi:10.3390/ijms222010912.
- Zeve D, Stas E, de Sousa Casal J, et al. Robust differentiation of human enteroendocrine cells from intestinal stem cells. Nat Commun. 2022;13(1):261. doi:10.1038/s41467-021-27901-5.
- Courtney CM, Onufer EJ, Seiler KM, Warner BW. An anatomic approach to understanding mechanisms of intestinal adaptation. Semin Pediatr Surg. 2018;27(4):229–236. doi:10.1053/j.sempedsurg.2018.07.00.
- Drozdowski LA, Clandinin T, Thomson ABR. Ontogeny, growth and development of the small intestine: understanding pediatric gastroenterology. World J Gastroenterol. 2010;16(7):787–799. doi:10.3748/wjg.v16.i7.787.
- Rao JN, Xiao L, Wang J-Y. Polyamines in gut epithelial renewal and barrier function. Physiology (Bethesda). 2020;35(5):328–337. doi:10.1152/physiol.00011.2020.
- Xiao L, Rao JN, Wang J-Y. RNA-binding proteins and long noncoding RNAs in intestinal epithelial autophagy and barrier function. Tissue Barriers. 2021;9(2):1895648. doi:10.1080/21688370.2021.1895648.
- Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340(6137):1190–1194. doi:10.1126/science.1234852.
- Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol. 2021;22(1):39–53. doi:10.1038/s41580-020-0278-0.
- Dudhwala ZM, Hammond PD, Howarth GS, Cummins AG. Intestinal stem cells promote crypt fission during postnatal growth of the small intestine. BMJ Open Gastroenterol. 2020;7(1):e000388. doi:10.1136/bmjgast-2020-000388.
- Tappenden KA. Intestinal adaptation following resection. JPEN J Parenter Enteral Nutr. 2014;38(1 Suppl):23S–31S. doi:10.1177/0148607114525210.
- Tai CC, Curtis J, Sala F, et al. Induction of fibroblast growth factor 10 (FGF10) in the ileal crypt epithelium after massive small bowel resection suggests a role for FGF10 in gut adaptation. Dev Dyn. 2009;238(2):294–301. doi:10.1002/dvdy.21667.
- Otterburn DM, Arthur LG, Timmapuri SJ, et al. Proteasome gene upregulation: a possible mechanism for intestinal adaptation. J Pediatr Surg. 2005;40(2):377–380. doi:10.1016/j.jpedsurg.2004.10.024.
- Washizawa N, Gu L, Openo K, et al. Comparative effects of glucagon-like peptide-2 (GLP-2), growth hormone (GH), and keratinocyte growth factor (KGF) on markers of gut adaptation after massive small bowel resection in rats. JPEN J Parenter Enteral Nutr. 2004;28(6):399–409. doi:10.1177/0148607104028006399.
- Balakrishnan A. Micromanaging the gut: unravelling the regulatory pathways that mediate the intestinal adaptive response. Ann R Coll Surg Engl. 2018;100(3):165–171. doi:10.1308/rcsann.2017.0174.
- Erwin CR, Falcone J, Stern LE, et al. Analysis of intestinal adaptation gene expression by cDNA expression arrays. JPEN J Parenter Enteral Nutr. 2000;24(6):311–316. doi:10.1177/0148607100024006311.
- Martin GR, Beck PL, Sigalet DL. Gut hormones, and short bowel syndrome: the enigmatic role of glucagon-like peptide-2 in the regulation of intestinal adaptation. World J Gastroenterol. 2006;12(26):4117–4129. doi:10.3748/wjg.v12.i26.4117.
- Drucker DJ. Glucagon-like peptide 2. J Clin Endocrinol Metab. 2001;86(4):1759–1764. doi:10.1210/jcem.86.4.7386.
- Le Roux CW, Borg C, Katharina W, et al. Gut hypertrophy after gastric bypass is associated with increased glucagon-like peptide 2 and intestinal crypt cell proliferation. Ann Surg. 2010;252(1):50–56. doi:10.1097/SLA.0b013e3181d3d21f.
- Wildhaber BE, Lynn KN, Yang H, Teitelbaum DH. Total parenteral nutrition-induced apoptosis in mouse intestinal epithelium: regulation by the Bcl-2 protein family. Pediatr Surg Int. 2002;18(7):570–575. doi:10.1007/s00383-002-0869-1.
- Pöyhönen M, Takala J, Pitkänen O, et al. Urinary excretion of polyamines in patients with surgical and accidental trauma: effect of total parenteral nutrition. Metabolism. 1997;42(1):44–51. doi:10.1016/0026-0495(93)90170-s.
- Chung HK, Xiao L, Jaladanki KC, Wang J-Y. Regulation of paneth cell function by RNA-binding proteins and noncoding RNAs. Cells. 2021;10(8):2107. doi:10.3390/cells10082107.
- Balakrishnan A, Stearns A, Park P, et al. Upregulation of proapoptotic microRNA mir-125a after massive small bowel resection in rats. Ann Surg. 2012;255(4):747–753. doi:10.1097/SLA.0b013e31824b485a.
- Bajinting A, Seiler K, Kanke M, et al. Su1091 – subepithelial microRNA expression during adaptation to massive small bowel resection. Gastroenterology. 2019;156(6):S-1436–S-1437. doi:10.1016/S0016-5085(19)40642-2.
- Langi G, Szczerbinski L, Kretowski A. Meta-analysis of differential miRNA expression after bariatric surgery. J Clin Med. 2019;8(8):1220. doi:10.3390/jcm8081220.
- Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282(32):23716–23724. doi:10.1074/jbc.M701805200.
- Xiao L, Cui Y-H, Rao JN, et al. Regulation of cyclin-dependent kinase 4 translation through CUG-binding protein 1 and microRNA-222 by polyamines. Mol Biol Cell. 2011;22(17):3055–3069. doi:10.1091/mbc.E11-01-0069.
- Jiang L-P, Wang S, Chung HK, et al. MiR-222 represses expression of zipcode binding protein-1 and phospholipase c-γ1 in intestinal epithelial cells. Am J Physiol Cell Physiol. 2019;316(3):C415–C423. doi:10.1152/ajpcell.00165.2018.
- Chung HK, Chen Y, Rao JN, et al. Transgenic expression of miR-222 disrupts intestinal epithelial regeneration by targeting multiple genes including Frizzled-7. Mol Med. 2015;21(1):676–687. doi:10.2119/molmed.2015.00147.
- Yan B, Guo Q, Fu F-j, et al. The role of miR-29b in cancer: regulation, function, and signaling. Onco Targets Ther. 2015;8:539–548. doi:10.2147/OTT.S75899.
- Wang H, Guan X, Tu Y, et al. MicroRNA-29b attenuates non-small cell lung cancer metastasis by targeting matrix metalloproteinase 2 and PTEN. J Exp Clin Cancer Res. 2015;34(1):59. doi:10.1186/s13046-015-0169-y.
- Ma F, Xu S, Liu X, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol. 2011;12(9):861–869. doi:10.1038/ni.2073.
- Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therap Adv Gastroenterol. 2015;8(1):4–22. doi:10.1177/1756283X14547360.
- Xiao L, Rao JN, Zou T, et al. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell. 2013;24(19):3038–3046. doi:10.1091/mbc.e13-05-0287.
- Zou T, Rao JN, Liu L, et al. JunD enhances miR-29b levels transcriptionally and posttranscriptionally to inhibit proliferation of intestinal epithelial cells. Am J Physiol Cell Physiol. 2015;308(10):C813–C824. doi:10.1152/ajpcell.00027.2015.
- Wang J-Y, Cui Y-H, Xiao L, et al. Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol. 2018;38(13):e00010-18. doi:10.1128/MCB.00010-18.
- Li Y, Chen G, Wang J-Y, et al. Post-transcriptional regulation of Wnt co-receptor LRP6 and RNA-binding protein HuR by miR-29b in intestinal epithelial cells. Biochem J. 2016;473(11):1641–1649. doi:10.1042/BCJ20160057.
- Ouyang M, Su W, Xiao L, et al. Modulation by miR-29b of intestinal epithelium homoeostasis through the repression of menin translation. Biochem J. 2015;465(2):315–323. doi:10.1042/BJ20141028.
- Zhuang R, Rao JN, Zou T, et al. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013;41(16):7905–7919. doi:10.1093/nar/gkt565.
- Qi J, Yu J-Y, Shcherbata HR, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8(22):3729–3741. doi:10.4161/cc.8.22.10033.
- Xu T, Zhu Y, Xiong Y, et al. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50(1):113–121. doi:10.1002/hep.22919.
- Bhattacharya A, Schmitz U, Wolkenhauer O, et al. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene. 2013;32(26):3175–3183. doi:10.1038/onc.2012.324.
- Zhang Y, Zhang Y, Xiao L, et al. Cooperative repression of insulin-like growth factor type 2 receptor translation by MicroRNA 195 and RNA-binding protein CUGBP1. Mol Cell Biol. 2017;37(19) doi:10.1128/MCB.00225-17.
- Wang SR, Rathor N, Kwon M, et al. miR-195 regulates intestinal epithelial restitution after wounding by altering actin-related protein-2 translation. Am J Physiol Cell Physiol. 2022;322(4):C712–C722. doi:10.1152/ajpcell.00001.2022.
- Xiao L, Wang J-Y. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr Opin Pharmacol. 2014;19:46–53. doi:10.1016/j.coph.2014.07.006.
- Tong Z, Liu N, Lin L, et al. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed Pharmacother. 2015;75:129–136. doi:10.1016/j.biopha.2015.07.036.
- Bernal NP, Stehr W, Coyle R, et al. Epidermal growth factor receptor signaling regulates Bax and Bcl-w expression and apoptotic responses during intestinal adaptation in mice. Gastroenterology. 2006;130(2):412–423. doi:10.1053/j.gastro.2005.11.006.
- Cheng W, Wang K, Zhao Z, et al. Exosomes-mediated transfer of miR-125a/b in cell-to-cell communication: a novel mechanism of genetic exchange in the intestinal microenvironment. Theranostics. 2020;10(17):7561–7580. doi:10.7150/thno.41802.
- Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–D144. doi:10.1093/nar/gkj112.
- Cao S, Xiao L, Rao JN, et al. Inhibition of Smurf2 translation by miR-322/503 modulates TGF-β/Smad2 signaling and intestinal epithelial homeostasis. Mol Biol Cell. 2014;25(8):1234–1243. doi:10.1091/mbc.e13-09-0560.
- He Y, Cai Y, Pai PM, et al. The causes and consequences of miR-503 dysregulation and its impact on cardiovascular disease and cancer. Front Pharmacol. 2021;12:629611. doi:10.3389/fphar.2021.629611.
- Cui Y-H, Xiao L, Rao JN, et al. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2012;23(1):151–162. doi:10.1091/mbc.E11-05-0456.
- Chassin C, Kocur M, Pott J, et al. MiR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 2010;8(4):358–368. doi:10.1016/j.chom.2010.09.005.
- MohanKumar K, Namachivayam K, Sivakumar N, et al. Severe neonatal anemia increases intestinal permeability by disrupting epithelial adherens junctions. Am J Physiol Gastrointest Liver Physiol. 2020;318(4):G705–G716. doi:10.1152/ajpgi.00324.2019.
- Yu T, Lu X-J, Shan T-D, et al. Overexpression of miR-429 impairs intestinal barrier function in diabetic mice by down-regulating occludin expression. Cell Tissue Res. 2016;366(2):341–352. doi:10.1007/s00441-016-2435-5.
- Shi C, Liang Y, Yang J, et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One. 2013;8(6):e66814. doi:10.1371/journal.pone.0066814.
- Zhou Q, Souba WW, Croce CM, Verne GN. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59(6):775–784. doi:10.1136/gut.2009.181834.
- Martínez C, Rodiño-Janeiro BK, Lobo B, et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut. 2017;66(9):1537–1538. doi:10.1136/gutjnl-2016-311477.
- Rawat M, Nighot M, Al-Sadi R, et al. IL1B increases intestinal tight junction permeability by up-regulation of MIR200C-3p, which degrades occludin mRNA. Gastroenterology. 2020;159(4):1375–1389. doi:10.1053/j.gastro.2020.06.038.
- Hou Q, Huang Y, Zhu S, et al. MiR-144 increases intestinal permeability in IBS-D rats by targeting OCLN and ZO1. Cell Physiol Biochem. 2017;44(6):2256–2268. doi:10.1159/000486059.
- Statello L, Guo C-J, Chen L-L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi:10.1038/s41580-020-00315-9.
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. doi:10.1038/ng.3192.
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi:10.1101/gr.132159.111.
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011.
- Bejerano G, Pheasant M, Makunin I, et al. Ultraconserved elements in the human genome. Science. 2004;304(5675):1321–1325. doi:10.1126/science.1098119.
- Xiao L, Wu J, Wang J-Y, et al. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology. 2018;154(3):599–611. doi:10.1053/j.gastro.2017.10.009.
- Kalakonda S, Xiao L, Chung HK, et al. 1015 - Novel long noncoding RNA UC.230 enhances growth of the intestinal epithelium by down-regulating microRNA 222. Gastroenterology. 2018;154(6):S-192. doi:10.1016/S0016-5085(18)31047-3.
- Ota T, Suzuki Y, Nishikawa T, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36(1):40–45. doi:10.1038/ng1285.
- Khaitan D, Dinger M, Mazar J, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71(11):3852–3862. doi:10.1158/0008-5472.CAN-10-4460.
- Li J, Chen Y, Chen Z, et al. SPRY4-IT1: a novel oncogenic long non-coding RNA in human cancers. Tumour Biol. 2017;39(6):1010428317711406. doi:10.1177/1010428317711406.
- Xiao L, Rao JN, Cao S, et al. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol Biol Cell. 2016;27(4):617–626. doi:10.1091/mbc.e15-10-0703.
- Wu B, Zhang Y, Yu Y, et al. Long noncoding RNA H19: a novel therapeutic target emerging in oncology via regulating oncogenic signaling pathways. Front Cell Dev Biol. 2021;9:796740. doi:10.3389/fcell.2021.796740.
- Geng H, Bu H-F, Liu F, et al. In inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology. 2018;155(1):144–155. doi:10.1053/j.gastro.2018.03.058.
- Yu T-X, Chung HK, Xiao L, et al. Long noncoding RNA H19 impairs the intestinal barrier by suppressing autophagy and lowering Paneth and goblet cell function. Cell Mol Gastroenterol Hepatol. 2020;9(4):611–625. doi:10.1016/j.jcmgh.2019.12.002.
- Zou T, Jaladanki SJ, Liu L, et al. H19 long non-coding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol. 2016;36(9):1332–1341. doi:10.1128/MCB.01030-15.
- Su Z, Zhi X, Zhang Q, et al. LncRNA H19 functions as a competing endogenous RNA to regulate AQP3 expression by sponging miR-874 in the intestinal barrier. FEBS Lett. 2016;590(9):1354–1364. doi:10.1002/1873-3468.12171.
- Zhu P, Wu J, Wang Y, et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 2018;20(10):1134–1144. doi:10.1038/s41556-018-0194-0.
- Chen T, Xue H, Lin R, Huang Z. MiR-34c and PlncRNA1 mediated the function of intestinal epithelial barrier by regulating tight junction proteins in inflammatory bowel disease. Biochem Biophys Res Commun. 2017;486(1):6–13. doi:10.1016/j.bbrc.2017.01.115.
- Ma D, Cao Y, Wang Z, et al. CCAT1 lncRNA promotes inflammatory bowel disease malignancy by destroying intestinal barrier via downregulating miR-185-3p. Inflamm Bowel Dis. 2019;25(5):862–874. doi:10.1093/ibd/izy381.
- Wu F, Huang Y, Dong F, Kwon JH. Ulcerative colitis-associated long noncoding RNA, BC012900, regulates intestinal epithelial cell apoptosis. Inflamm Bowel Dis. 2016;22(4):782–795. doi:10.1097/MIB.0000000000000691.
- Akıncılar SC, Wu L, Ng QF, et al. NAIL: An evolutionarily conserved lncRNA essential for licensing coordinated activation of p38 and NFκB in colitis. Gut. 2021;70(10):1857–1871. doi:10.1136/gutjnl-2020-322980.
- Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi:10.1038/nrg.2016.114.
- Zhou W-Y, Cai Z-R, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. doi:10.1186/s12943-020-01286-3.
- Liu C-X, Chen L-L. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185(12):2016–2034. doi:10.1016/j.cell.2022.04.021.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928.
- Li Y, Zheng F, Xiao X, et al. Circ-HIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi:10.15252/embr.201643581.
- Xiao L, Ma X-X, Luo J, et al. Circular RNA CircHIPK3 promotes homeostasis of the intestinal epithelium by reducing microRNA 29b function. Gastroenterology. 2021;161(4):1303–1317.e3. doi:10.1053/j.gastro.2021.05.060.
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7(1):11215. doi:10.1038/ncomms11215.
- Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi:10.1080/15476286.2017.1279788.
- Li X-X, Xiao L, Chung HK, et al. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol. 2020;40(6):e00492–19. doi:10.1128/MCB.00492-19.
- Chung HK, Xiao L, Han N, et al. Circular RNA Cdr1as inhibits proliferation and delays injury-induced regeneration of the intestinal epithelium. JCI Insight. 2024;e169716. doi:10.1172/jci.insight.169716.
- Gallo S, Kong E, Ferro I, Polacek N. Small but powerful: the human vault RNAs as multifaceted modulators of pro-survival characteristics and tumorigenesis. Cancers (Basel). 2022;14(11):2787. doi:10.3390/cancers14112787.
- Hahne JC, Lampis A, Valeri N. Vault RNAs: hidden gems in RNA and protein regulation. Cell Mol Life Sci. 2020;78(4):1487–1499. doi:10.1007/s00018-020-03675-9.
- Horos R, Büscher M, Kleinendorst R, et al. The small non-coding vault RNA1-1 acts as a riboregulator of autophagy. Cell. 2019;176(5):1054–1067.e12. doi:10.1016/j.cell.2019.01.030.
- Büscher M, Horos R, Hentze MW. ‘High vault-age’: non-coding RNA control of autophagy. Open Biol. 2020;10(2):190307. doi:10.1098/rsob.190307.
- Büscher M, Horos R, Huppertz I, et al. Vault RNA1–1 riboregulates the autophagic function of p62 by binding to lysine 7 and arginine 21, both of which are critical for p62 oligomerization. RNA. 2022;28(5):742–755. doi:10.1261/rna.079129.122.
- Wallis K, Walters JR, Gabe S. Short bowel syndrome: the role of GLP-2 on improving outcome. Curr Opin Clin Nutr Metab Care. 2009;12(5):526–532. doi:10.1097/MCO.0b013e32832d23cd.
- Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics—challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629–651. doi:10.1038/s41573-021-00219-z.
- Huang CK, Kafert-Kasting S, Thum T. Preclinical and clinical development of noncoding RNA therapeutics for cardiovascular disease. Circ Res. 2020;126(5):663–678. doi:10.1161/CIRCRESAHA.119.315856.
- Shinn J, Lee K, Lee SA, et al. Oral nanomedicines for siRNA delivery to treat inflammatory bowel disease. Pharmaceutics. 2022;14(9):1969. doi:10.3390/pharmaceutics14091969.

