Abstract
Stevioside and rebaudioside A are the chief diterpene glycosides present in the leaves of Stevia rebaudiana. Rebaudioside A imparts a desirable sweet taste, while stevioside produces a residual bitter aftertaste. Enzymatic synthesis of rebaudioside A from stevioside can increase the ratio of rebaudioside A to stevioside in steviol glycoside products, providing a conceivable strategy to improve the organoleptic properties of steviol glycoside products. Here, we demonstrate the efficient conversion of stevioside to rebaudioside A by coupling the activities of recombinant UDP-glucosyltransferase UGT76G1 from S. rebaudiana and sucrose synthase AtSUS1 from Arabidopsis thaliana. The conversion occurred via regeneration of UDP-glucose by AtSUS1. UDP was applicable as the initial material instead of UDP-glucose for UDP-glucose recycling. The amount of UDP could be greatly reduced in the reaction mixture. Rebaudioside A yield in 30 h with 2.4 mM stevioside, 7.2 mM sucrose, and 0.006 mM UDP was 78%.
Graphical abstract
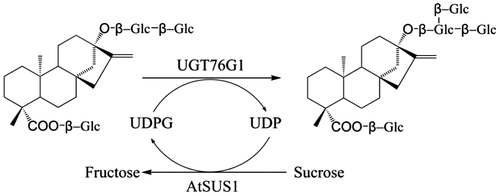
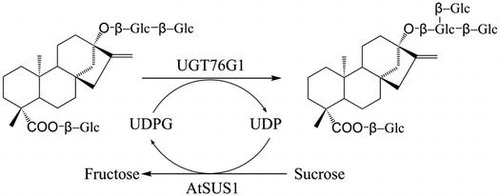
Concept of the two-enzyme system for efficient synthesis of rebaudioside A from stevioside. UGT76G1, UDP-glucosyltransferase from Stevia rebaudiana; AtSUS1, sucrose synthase from Arabidopsis thaliana.
Steviol glycosides are a group of sweet-tasting compounds extracted and purified from the leaves of Stevia rebaudiana Bertoni. More than 30 steviol glycosides have been discovered in S. rebaudiana, of which stevioside and rebaudioside A are the predominant sweetening agents.Citation1,2) Their concentrations vary widely depending on the genotype, cultivation conditions, and harvesting time of the plant.Citation3) The typical proportion of stevioside and rebaudioside A in dry leaf matter of wild stevia plants is 9.1 and 3.8%, respectively.Citation4) Great attention has been paid to steviol glycosides for their utility as low-cost natural sweeteners in food and beveragesCitation5) because of the following: their high-intensity sweetness; low calorie count; non-nutritive nature; high stability;Citation6,7) and reported therapeutic properties such as antihyperglycemic, antihypertensive, anti-inflammatory, antitumor, antidiarrheal, diuretic, and immunomodulatory effects.Citation8,9) The Joint FAO/WHO Expert Committee on Food Additives has established a monograph for steviol glycosides, and highly pure steviol glycosides have been officially approved as food additives in the United States and European Union. The United States Food and Drug Administration granted “generally recognized as safe” regulatory acceptance of rebaudioside A in 2008.Citation10)
Usually, stevioside and rebaudioside A coexist in commercially available steviol glycoside products. Stevioside is 250–300 times sweeter than sucrose but produces a bitter aftertaste, which constrains its use for human consumption and application in food and pharmaceutical products.Citation11–13) Rebaudioside A is sweeter than stevioside and produces a clean, sweet taste with no significant undesirable aftertaste characteristics.Citation14) For debittering steviosides, stevioside modification catalyzed by α-amylaseCitation15) or β-cyclodextrin glucanotransferase has been applied.Citation16–18) In this study, we propose a strategy that increases the rebaudioside A/stevioside ratio in stevia extracts by the conversion of stevioside to rebaudioside A, thereby enabling a low-cost preparation of high-purity rebaudioside A (Fig. ). In vitro glycosylation of stevioside to produce rebaudioside A was catalyzed by the Stevia UDP-glucosyltransferase UGT76G1, of which the glucosyltransferase activity for specific generation of rebaudioside A from stevioside was previously confirmed, with UDP-glucose as the activated sugar donor.Citation19,20) The UDP-glucose availability for this reaction was facilitated by the activity of a sucrose synthase from Arabidopsis thaliana (AtSUS1), which catalyzes the generation of UDP-glucose and fructose from sucrose and UDP, respectively. UDP-glucose and UDP were investigated as initial materials in the two-enzyme system.
Materials and methods
Synthesis of A. thaliana sucrose synthase gene
The coding region of AtSUS1 cDNA (GenBank accession no: NM122090), with codons optimized for expression in Saccharomyces cerevisiae in the earlier study, was synthesized using a two-step PCR-based method.Citation21)
The first step was the PCR-based synthesis of ~600-bp (base pair) fragments using a high-fidelity PrimeSTAR HS DNA polymerase (Takara, Dalian, China) from 72 oligonucleotides of 59 nt (nucleotide) each, with 18–29-nt overlaps at both 5′ and 3′ ends between adjacent oligonucleotides. The oligonucleotide fragments were designed using the specialized oligonucleotide design program that is DNAWorks.Citation22) The melting temperature (Tm) of the overlapping regions was kept similar, and in the gene assembly experiment, a Tm of 62 °C was found to be most suitable according to the DNAWorks. The oligonucleotides were diluted to a concentration of 10 μM with double-distilled water for later use. The 72 oligonucleotides were divided into four groups, and equal volumes of oligonucleotides in each group were mixed and used as the inner oligonucleotides. Assembly of PCR was carried out in a 50-μL reaction mixture containing 1 pmol of each inner oligonucleotide, 20 pmol of each outer oligonucleotide, 1× PCR buffer, 4 μL dNTP (2.5 mM each), and 1.25 U of PrimeSTAR HS DNA polymerase. The amplification was comprised of 30 cycles at 98 °C for 10 s, followed by 68 °C for 1 min.
The second step was assembling the full-length gene by overlap-extension PCR. Four microliters of the mixture combined from four different products in the first-step PCRs was used as the template for the second PCR, with 2 μL of the two outermost oligonucleotides as primers in a final volume of 50 μL. The PCR protocol was the same except for annealing conditions, i.e., 68 °C for 2 min 30 s. A 3′ overhang was added to the final PCR product by incubation with additional 1 μL dATP (10 mM) and 0.5 μL Taq DNA polymerase (5 U/μL) (Takara) at 72 °C for another 30 min. The product was subsequently purified by a gel extraction kit (Takara). The purified DNA fragment was then cloned into a pMD®18-T vector (Takara). Several positive clones were sequenced by a contract research organization, GenScript (Nanjing, China), to check for fidelity.
The above synthesized sucrose synthase gene was used to express in Escherichia coli in this study. To avoid the rare codon effect on the recombinant protein expression, E. coli Rosetta (DE3) (Novagen), which belongs to BL21 derivatives designed to enhance the expression of eukaryotic proteins that contain codons rarely used in E. coli, was used as the host strain.
Plasmid construction
The codon-optimized gene encoding UGT76G1 in Stevia (DDBJ accession number: LC037193), synthesized by GenScript, was previously cloned into the expression vector pET-28a(+) (Novagen), yielding plasmid pEUGT. The primers P1 (GGAATTCCATATGGAAAATAAGACTGAAACTACTG) and P2 (CCGCTCGAGTTATAATGATGAAATATAAGAAACCAA) (Nde I and Xho I restriction endonuclease sites underlined) were used for pEUGT construction.
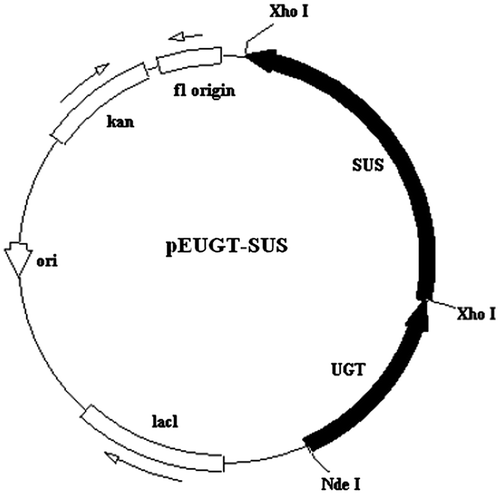
The 2427-bp modified sucrose synthase gene was subcloned into pEUGT using the CloneEZ® PCR cloning kit, with the primers P3 (TTTCATCATTATAACTCGAGATATACATATGGCGAATGCCGAGAGAATGAT) and P4 (GGTGGTGGTGCTCGAGTTAATCGTCCTGGGCTAATGGC). The Xho I restriction endonuclease sites were underlined, and the Shine-Dalgarno (SD) sequence was boxed. The resulting plasmid coexpressing UGT76G1 and AtSUS1 was designated as pEUGT-SUS (Fig. ).
Expression of recombinant glucosyltransferase and sucrose synthase in E. coli
Individual colonies of E. coli Rosetta (DE3) cells transformed with the plasmid pEUGT or pEUGT-SUS were grown in Luria Bertani medium containing 50 μg/mL kanamycin and 34 μg/mL chloramphenicol overnight at 37 °C with shaking at 200 rpm. Next, aliquots of the cultures were inoculated into 50 mL of fresh auto-induction medium (15 g/L tryptone, 25 g/L yeast extract, 10 g/L NaCl, 2 g/L glucose, and 3 g/L lactose) supplemented with antibiotics and incubated at 30 °C with shaking at 200 rpm for 12–13 h.
Enzymatic synthesis of rebaudioside A with UGT76G1 and AtSUS1
After pEUGT-SUS expression in E. coli, the cells were harvested and washed twice in potassium phosphate buffer (100 mM, pH 7.2). A lysate containing soluble proteins was obtained by sonication on an ice-water bath and subsequent clarification by centrifugation at 4 °C. The supernatant was used as the crude extract. Total protein concentrations were measured by the Bradford assay using bovine serum albumin (BSA) as a standard.Citation23) The substrate stevioside was incubated with aliquots of the crude extract at 30 °C in a reaction mixture containing varying concentrations of UDP-glucose or UDP, sucrose, and 3 mM MgCl2 in potassium phosphate buffer (100 mM, pH 7.2) for synthesis of rebaudioside A.
Glucosyltransferase activity assayCitation24)
The UGT76G1 expressed by E. coli was subjected to the enzyme assay reaction. The assay mixture contained 1 mg of total protein from the crude extract, 1 mM stevioside, 2 mM UDP-glucose, and 3 mM MgCl2 in potassium phosphate buffer (100 mM, pH 7.2), in a total volume of 5 mL. Reactions were performed at 30 °C for 1 h. The samples were taken and prepared for high-performance liquid chromatography (HPLC) analysis. One unit (U) of glucosyltransferase activity was defined as the amount of enzyme that produced 1 μmol of rebaudioside A per min under the described conditions.
As the two genes, respectively, coding UGT76G1 and AtSUS1 in the pEUGT-SUS construct were expressed as one Polycistron driven by the T7 promoter, the UDP-glucosyltransferase activity was defined to describe the amount of recombinant enzymes which were used in the above reaction mixtures.
HPLC analysisCitation3)
Samples of the reaction mixture described above were extracted three times with equal volumes of water-saturated 1-butanol and then dried using a nitrogen purge device (QYN100-1) (Joyn Electronic, Shanghai, China). The pooled butanol fractions were dried completely and re-dissolved in 300 μL water-saturated 1-butanol.
The analysis of rebaudioside A was conducted on an HPLC system (Agilent 1290 Infinity, Agilent Technologies) using a GalaksilTM BF-NH2 column (4.6 × 250 mm, 5 μm 120 Å) maintained at 40 °C with UV detection at 210 nm. The flow rate was 1 mL/min using 20% H2O in CH3CN (pH adjusted to 4.2 with acetic acid) as the mobile phase. The injection volume was 10 μL. Rebaudioside A standard (99%) for HPLC was obtained for Wako Pure Chemical Industries, Ltd (Japan). The rebaudioside A yield was calculated as following: rebaudioside A yield (%) = C(RA)/C(St). There C(St) represents the initial stevioside mole concentration; C(RA) represents the increased rebaudioside A mole concentration after the reaction.
HPLC-MS analysis
HPLC-MS analysis was conducted on an Agilent 6460 HPLC system, coupled with negative electrospray ionization (ESI) mass spectrometry (MS) method. Mass spectra in the negative ion mode were operated under the following conditions: fragmenter voltage = 130 eV, voltage = 3500 V, nebulizer pressure = 45 psi, capillary temperature = 325 °C, m/z range = 100–1200. The analytical column used was a GalaksilTM BF-NH2 column (4.6 × 250 mm, 5 μm 120 Å) maintained at 30 °C, and the flow rate was 0.6 mL/min using 30% H2O in CH3CN as the mobile phase. The injection volume was 20 μL.
Results and discussion
Heterologous expression of UGT76G1 and AtSUS1
The pattern of glycosylation heavily influences the sweetness and taste perception of a mixture of different steviol glycosides. The enzyme UGT76G1 from S. rebaudiana catalyzes the conversion of stevioside, the most abundant steviol glycoside, to rebaudioside A, which possesses superior organoleptic properties,Citation19,20,25) providing an approach to change the glycosylation patterns of steviol glycoside products and accordingly improve their taste quality.
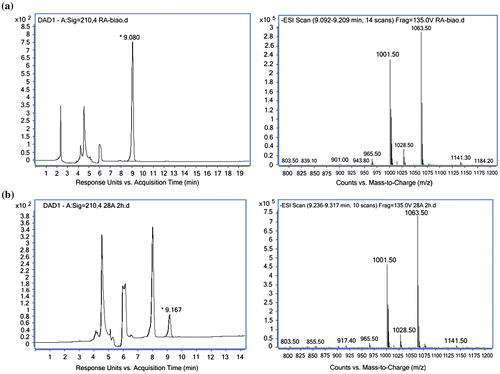
In our study, when the samples were taken immediately after incubation with 1.6 mU/mL enzymes from the crude extract, 1 mM stevioside and 2 mM UDP-glucose, 0.02 mM rebaudioside A was detected in the reaction mixture (Fig. (a)). After incubation for 2 h, the amount of rebaudioside A reached 0.28 mM (Fig. (b)). HPLC-MS analysis of the product ion scan spectrum was carried out, with rebaudioside A standard similarly prepared as control. The characteristic fragmentation of [M−H]− was identical with rebaudioside A standard that were at m/z 965.5 (Fig. ). Therefore, the product generated in the reaction mixture was confirmed as rebaudioside A. These results indicate that the recombinant UGT76G1 coded by the synthetic gene was functional when expressed in E. coli.
Fig. 3. HPLC showing the peaks of stevioside and rebaudioside A detected in the glucosyltransferase activity assay.
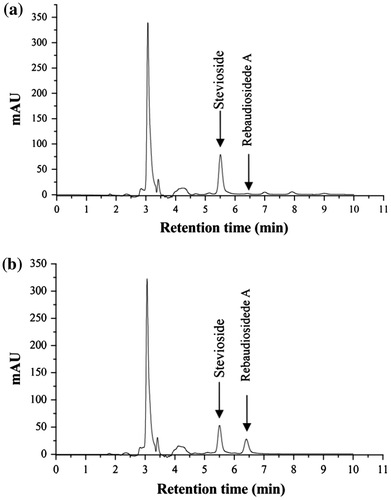
Fig. 4. HPLC-MS analysis of rebaudioside A standard (a) and the sample prepared for the glucosyltransferase activity assay (b).
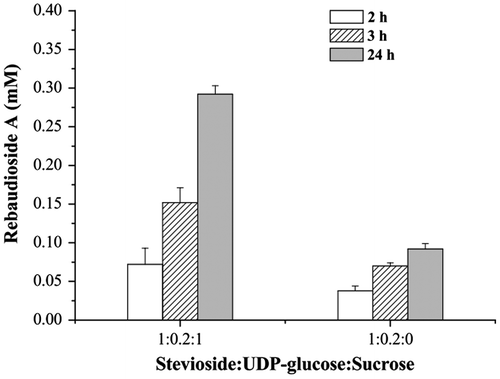
Generally, enzymatic glycosylation is not scalable because addition of the activated sugar donor is quite expensive. To reduce the cost of supplying of UDP-glucose in such reactions, in situ regeneration of UDP-glucose is an advantageous option.Citation26,27) Here, recombinant E. coli containing pEUGT-SUS was induced and crude extracts (2 mU/mL enzymes) from the E. coli cells expressing UGT76G1 and AtSUS1 were added to the reaction mixtures, where the concentrations of stevioside was 0.6 mM, and the initial stevioside:UDP-glucose:sucrose mole ratios were 1:0.2:1 and 1:0.2:0, respectively (Fig. ). After incubation for 24 h, the concentration of rebaudioside A was as high as 0.29 mM with sucrose, which was 3.2 folds of that without sucrose (0.09 mM, with a rebaudioside A yield of 15.3%). This result indicates that UDP-glucose was generated from the sucrose cleavage when the activities of AtSUS1 and UGT76G1 were combined.
Effect of the initial sugar donor on rebaudioside A synthesis
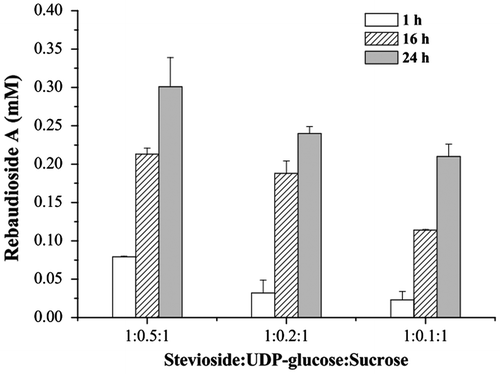
Although enzymatic glycosylation of stevioside to rebaudioside A using the recombinant UGT76G1 was shown to be efficient, the process still required UDP-glucose. By coupling the activity of sucrose synthase, UDP-glucose initially added to the reaction mixture could be reduced to one-tenth of the normal level, and the yields were 19% after 16 h and 35% after 24 h (Fig. ). With a higher UDP-glucose concentration in the system, more rebaudioside A was obtained in the same reaction time. In the reaction mixture with an initial stevioside:UDP-glucose:sucrose mole ratio of 1:0.5:1, the yield was as high as 50%, and the concentration of rebaudioside A was 0.30 mM after reaction by time of 24 h. But less cycle numbers of UDP-glucose was obtained at the high initial concentration of UDP-glucose, probably because UDP produced by the enzymatic glycosylation reaction was a potent inhibitor of glucosyltransferase activity.Citation28)
Fig. 6. Effect of the initial UDP-glucose concentrations on enzymatic production of rebaudioside A.
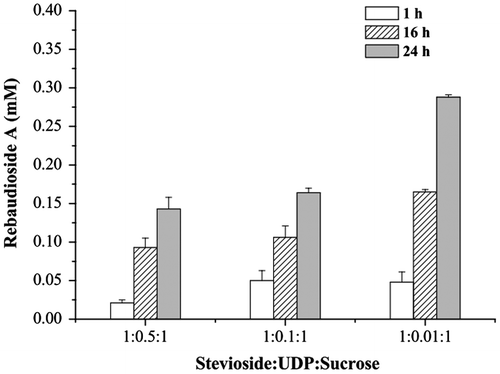
When UDP was added for recycling UDP-glucose in the reaction mixture, the inhibition resulting from high UDP concentration was prominent when comparing the systems with the initial stevioside:UDP:sucrose mole ratios of 1:0.5:1 and 1:0.01:1 (Fig. ). In the mixture with 1% UDP (0.006 mM), the amount of rebaudioside A was 0.29 mM, corresponding to the highest yield of 48% among the three compared systems after reaction by time of 24 h. The results indicate that inhibition of glycosylation reaction by UDP was alleviated by low concentration of UDP.
Effect of the initial sucrose concentration on rebaudioside A synthesis
The biological function of sucrose synthase (E.C. 2.4.2.13) is considered to be the synthesis of nucleotide diphosphate derivatives of glucose in the presence of high concentrations of sucrose, although its catalysis of both synthesis and cleavage of sucrose is freely reversible in vitro.Citation29) In our study, the reactions were carried out with 19.8 mU/mL enzymes from the crude extract, 0.6 mM stevioside, 0.006 mM UDP, and different concentrations of sucrose. Higher sucrose concentrations indeed resulted in a higher rebaudioside A yield in 20 h (Fig. ). As a result, the maximum yield of rebaudioside A reached 92% (0.55 mM) after reaction for 20 h, when the mole ratio of sucrose to steviosdie was 4:1. But the accumulation of rebaudioside A in the reaction mixture was close to that with the sucrose to stevioside ratio in 3:1. More than 25% concentration difference of rebaudioside A was found between the samples in the ratio of sucrose to stevioside 2:1 and 1:1.
Fig. 8. Effect of sucrose concentrations on enzymatic production of rebaudioside A.
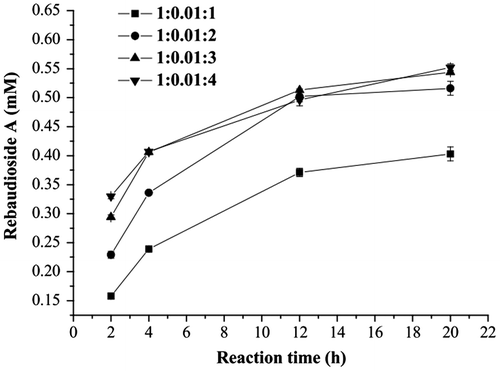
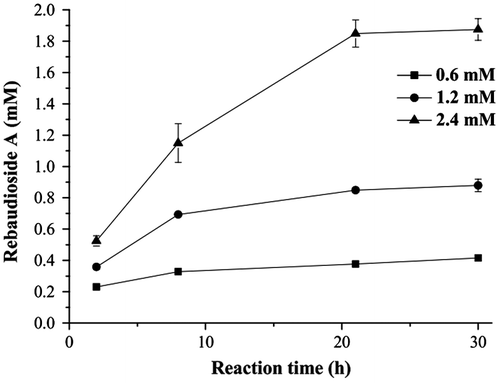
When the mole ratio of sucrose to stevioside was fixed at 3:1 and the initial UDP concentration was 0.006 mM, higher yields were reached with lower initial stevioside concentrations in 2 h. However, the rebaudioside A amount detected in the reaction mixture was 1.88 mM from 2.4 mM stevioside after reaction for 30 h, with the highest yield (78%), compared with that from 1.2 mM (73%) and 0.6 mM stevioside (70%) under the same conditions (Fig. ). The cycle numbers of UDP were approximately 70, 147, and 313 for the reaction system with 0.6, 1.2, and 2.4 mM of stevioside, respectively.
Fig. 9. Effect of stevioside concentrations on enzymatic production of rebaudioside A.
Since we used crude extract from cells expressing UGT76G1 and AtSUS1, the inclusion of traces of endogenous UDP and UDP-glucose in the reaction mixtures was unavoidable. However, combining such an in situ system for regeneration of UDP-glucose with glucosyl transfer catalyzed by stevia UDP-glucosyltransferase was investigated in this study for in vitro glycosylation of stevioside into rebaudioside A. Purification and immobilization of the UGT76G1 and AtSUS1 can probably increase the production efficiency, which will outline the course of future work.
Conclusion
UDP-glucosyltransferase UGT76G1 from S. rebaudiana and sucrose synthase AtSUS1 from A. thaliana were coexpressed in E. coli. Coupling the activities of these two recombinant enzymes resulted in the efficient synthesis of rebaudioside A from stevioside. It is noteworthy that the recombinant sucrose synthase AtSUS1 produced high levels of UDP-glucose so that a small amount of UDP could be used instead of UDP-glucose to initiate the reaction, which can considerably increase the cost effectiveness of the system.
Author contributions
Y.L. and M.Y. conceived and designed the study. Y.W., L.L.C., and Y.Y.L. performed the experiments and analyzed the data. Y.W. and Y.L. wrote the manuscript. M.Y., K.Q.C., and N.H. reviewed and commented on the manuscript. Y.L. and L.X. supported in the finance. All authors have read and approved of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was financially supported by the National High Technology Research and Development Program of China [grant number 2012AA022101], NSFC [grant number 21106068], Natural Science Foundation of Jiangshu Province of China [grant number BK2011801], Doctoral Fund of Ministry of Education of China [grant number 20113221120002], and Natural Science Foundation of the Higher Education Institutions of Jiangsu Province, China [grant number 10KJB530004].
References
- Wölwer-Rieck U. The leaves of Stevia rebaudiana (Bertoni), their constituents and the analyses thereof: a review. J. Agric. Food Chem. 2012;60:886–895.10.1021/jf2044907
- Ibrahim MA, Rodenburg DL, Alves K, et al. Minor diterpene glycosides from the leaves of stevia rebaudiana. J. Nat. Prod. 2014;77:1231–1235.10.1021/np4009656
- Kolb N, Herrera JL, Ferreyra DJ, Uliana RF. Analysis of sweet diterpene glycosides from Stevia rebaudiana: improved HPLC Method. J. Agric. Food Chem. 2001;49:4538–4541.10.1021/jf010475p
- Brandle JE, Starratt AN, Gijzen M. Stevia rebaudiana: its agricultural, biological, and chemical properties. Can. J. Plant Sci. 1998;78:527–536.10.4141/P97-114
- Ceunen S, Geuns JM. Steviol glycosides: chemical diversity, metabolism, and function. J. Nat. Prod. 2013;76:1201–1228.10.1021/np400203b
- Clos JF, DuBois GE, Prakash I. Photostability of rebaudioside A and stevioside in beverages. J. Agric. Food Chem. 2008;56:8507–8513.10.1021/jf801343e
- Wölwer-Rieck U, Tomberg W, Wawrzun A. Investigations on the stability of stevioside and rebaudioside A in soft drinks. J. Agric. Food Chem. 2010;58:12216–12220.10.1021/jf102894v
- Goyal SK, Samsher, Goyal RK. Stevia (Stevia rebaudiana) a bio-sweetener: a review. Int. J. Food Sci. Nutr. 2010;61:1–10.10.3109/09637480903193049
- Chatsudthipong V, Muanprasat C. Stevioside and related compounds: therapeutic benefits beyond sweetness. Pharmacol Ther. 2009;121:41–54.10.1016/j.pharmthera.2008.09.007
- Andress S. U.S. Food and Drug Administration, Agency Response Letter GRAS Notice No. GRN 000252, CFSAN/Office of Food Additive Safety. Whole Earth Sweetener Company LLC: Chicago, IL (2008).
- Brandle JE, Starratt AN, Gijzen M. Stevia rebaudiana: its agricultural, biological, and chemical properties. Can. J. Plant Sci. 1998;78:527–536.10.4141/P97-114
- Carakostas MC, Curry LL, Boileau AC, Brusick DJ. Overview: the history, technical function and safety of rebaudioside A, a naturally occurring steviol glycoside, for use in food and beverages. Food Chem. Toxicol. 2008;46:S1–S10.10.1016/j.fct.2008.05.003
- Scipioni GP, Ferreyra DJ, Acuña MG, Schmalko ME. Rebaudioside A release from matrices used in a yerba maté infusion. J. Food Eng. 2010;100:627–633.10.1016/j.jfoodeng.2010.05.011
- Prakash I, DuBois GE, Clos JF, Wilkens KL, Fosdick LE. Development of rebiana, a natural, non-caloric sweetener. Food Chem. Toxicol. 2008;46:S75–S82.10.1016/j.fct.2008.05.004
- Ye F, Yang R, Hua et al. Modification of stevioside using transglucosylation activity of Bacillus amyloliquefaciens α-amylase to reduce its bitter aftertaste. LWT – Food Sci. Technol. 2013;51:524–530.10.1016/j.lwt.2012.12.005
- Abelyan VA, Balayan AM, Ghochikyan VT, Markosyan AA. Transglycosylation of stevioside by cyclodextrin glucanotransferases of various groups of microorganisms. Appl. Biochem. Microbiol. 2004;40:129–134.10.1023/B:ABIM.0000018914.08571.50
- Jaitak V, Kaul VK, Bandna, et al. Simple and efficient enzymatic transglycosylation of stevioside by β-cyclodextrin glucanotransferase from Bacillus firmus. Biotechnol. Lett. 2009;31:1415–1420.10.1007/s10529-009-0020-7
- Li S, Li W, Xiao Q, Xia Y. Transglycosylation of stevioside to improve the edulcorant quality by lower substitution using cornstarch hydrolyzate and CGTase. Food Chem. 2013;138:2064–2069.10.1016/j.foodchem.2012.10.124
- Humphrey TV, Richman AS, Menassa R, Brandle JE. Spatial organisation of four enzymes from Stevia rebaudiana that are Involved in steviol glycoside synthesis. Plant Mol. Biol. 2006;61:47–62.10.1007/s11103-005-5966-9
- Richman A, Swanson A, Humphrey T, et al. Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major sweet glucosides of Stevia rebaudiana. Plant J. 2005;41:56–67.
- Xiong AS, Yao QH, Peng RH, et al. A simple, rapid, high-fidelity and cost-effective PCR-based two-step DNA synthesis method for long gene sequences. Nucleic Acids Res. 2004;32:e98.10.1093/nar/gnh094
- Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43.10.1093/nar/30.10.e43
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254.10.1016/0003-2697(76)90527-3
- Madhav H, Bhasker S, Chinnamma M. Functional and structural variation of uridine diphosphate glycosyltransferase (UGT) gene of Stevia rebaudiana-UGTSr involved in the synthesis of rebaudioside A. Plant Physiol. Biochem. 2013;63:245–253.10.1016/j.plaphy.2012.11.029
- Brandle JE, Telmer PG. Steviol glycoside biosynthesis. Phytochemistry. 2007;68:1855–1863.10.1016/j.phytochem.2007.02.010
- De Carvalho CC. Enzymatic and whole cell catalysis: finding new strategies for old processes. Biotechnol. Adv. 2011;29:75–83.10.1016/j.biotechadv.2010.09.001
- Tsai TI, Lee HY, Chang SH, et al. Effective sugar nucleotide regeneration for the large-scale enzymatic synthesis of Globo H and SSEA4. J. Am. Chem. Soc. 2013;135:14831–14839.10.1021/ja4075584
- Terasaka K, Mizutani Y, Nagatsu A, Mizukami H. In situ UDP-glucose regeneration unravels diverse functions of plant secondary product glycosyltransferases. FEBS Lett. 2012;586:4344–4350.10.1016/j.febslet.2012.10.045
- Figueroa CM, Asención Diez MD, Kuhn ML, et al. The unique nucleotide specificity of the sucrose synthase from Thermosynechococcus elongatus. FEBS Lett. 2013;587:165–169.10.1016/j.febslet.2012.11.011