ABSTRACT
Alginate is an acidic heteropolysaccharide produced by brown seaweed and certain kinds of bacteria. The cells of Sphingomonas sp. strain A1, a gram-negative bacterium, have several alginate-degrading enzymes in their cytoplasm and efficiently utilize this polymer for their growth. Sphingomonas sp. strain A1 cells can directly incorporate alginate into their cytoplasm through a transport system consisting of a “pit” on their cell surface, substrate-binding proteins in their periplasm, and an ATP-binding cassette transporter in their inner membrane. This review deals with the structural and functional aspects of bacterial systems necessary for the recognition and uptake of alginate.
Graphical Abstract
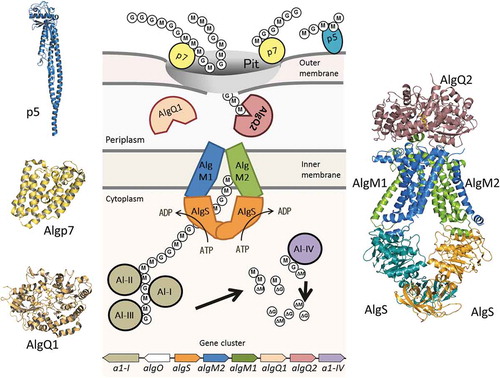
An overall picture of alginate import and depolymerization systems of Sphingomonas sp. strain A1.
Alginate is a linear acidic polysaccharide composed of β-D-mannuronate (M) and its C-5 epimer, α-L-guluronate (G) [Citation1]. It is widely distributed in the cell walls of brown seaweeds and is produced by certain bacteria such as Pseudomonas aeruginosa and Azotobacter vinelandii. Seaweed-derived alginate is widely used in a variety of fields, for example, in the food industry as a thickener and stabilizing agent, and in medicine as a covering agent for wounds and in surgical suture materials [Citation2,Citation3]. On the other hand, P. aeruginosa can form biofilms by using extra cellular partially-acetylated alginate [Citation4–Citation6], which makes infectious disease therapy for this bacterium difficult. In recent years, biofuel production using marine biomass has become an active field of research, and alginate from seaweeds has attracted attention as a raw material for ethanol production [Citation7,Citation8].
Sphingomonads are gram-negative rod-shaped bacteria that typically produce yellow-pigmented colonies and, uniquely among gram-negative bacteria, lack lipopolysaccharides, containing glycosphingolipids in their outer membrane instead [Citation9,Citation10]. Sphingomonas sp. strain A1 (strain A1) was isolated from soil and is a potent producer of alginate lyase [Citation11]. The strain A1 cell surface is covered with many large plaits, and a mouth-like pit is formed on their cell surface when grown on alginate-containing medium [Citation11,Citation12]. Alginate is found to be concentrated around this pit. In general, bacterial cells release enzymes on assimilating macromolecules including alginate and uptake depolymerized macromolecules into the cell. However, alginate uptake system in strain A1 is unique in that alginate-degrading enzymes are localized in the cytoplasm [Citation13], indicating that intact alginate polymers must be incorporated into the cytoplasm, which implies the presence of a unique system for the polymer uptake (). Some alginate-binding proteins are located on the surface of strain A1 cells. Examples include p5 and p7 (currently called Algp7), which were found by proteomic analysis of strain A1 outer membranes, and are thought to function in the recognition and concentration of alginate [Citation14]. In the periplasm, two solute-binding proteins, AlgQ1 and AlgQ2, bind alginate and pass it to a transporter, AlgM1M2SS, localized in the cytoplasmic membrane. Once alginate has been transferred to the cytoplasm, it is subsequently degraded by several alginate lyases to an unsaturated monosaccharide. Because M and G are C-5 epimers of each other, unsaturated sugars derived from M and G (ΔM and ΔG) are identical. They have C = C double bonds between their C-4 and C-5 atoms, and thus the difference in orientation of the carboxyl group is eliminated.
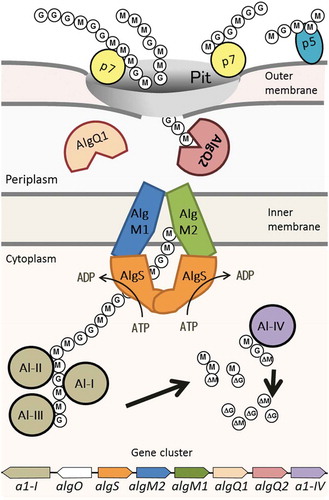
Figure 1. Alginate uptake in Sphingomonas sp. strain A1. An overall picture of alginate import and depolymerization systems. M and G represent the β-D-mannurorate and α-L-guluronate residues of alginate, respectively. AlgQ1 and AlgQ2 are solute-binding proteins that pass alginate to the ABC transporter. AlgM1, AlgM2, and AlgS form a heterotetramer (ABC transporter, AlgM1M2SS) and transport alginate across the inner membrane. A1-I to III are endo-type alginate lyases and A1-IV is an exo-type lyase. A1-I is divided to A1-II and A1-III. A series of actions by these lyases produce unsaturated monosaccharides (ΔM and ΔG), which are structurally the same. All of these proteins, together with a transcriptional regulator, AlgO, are coded by genes in the alginate-related gene cluster.
The alginate transporter localized in the cytoplasmic membrane of strain A1 is a member of an ATP-binding cassette (ABC) transporter that, when coupled to the hydrolysis of ATP, can translocate a wide variety of substrates across the membrane [Citation15,Citation16]. These transporters form a very large protein superfamily sharing similar structural features, and are present in all living organisms [Citation17–Citation21]. They consist of two homologous transmembrane domains and two nucleotide-binding domains. Bacterial ABC importers involved in the uptake of nutrients require additional periplasmic or lipid-anchored extracellular solute-binding proteins specific to a particular substrate [Citation22]. Type I, II, and III ABC importers have been identified, based on the folds of their transmembrane domains and other biochemical aspects [Citation23]. Type I ABC importers possess five to eight transmembrane helices in their transmembrane domain, while type II ABC importers have ten. Type III importers (also known as energy coupling-factor transporters) on the other hand, require a membrane-embedded component instead of a solute-binding protein. The overall structure of the type I ABC importer was first determined in the molybdate transporter [Citation24]. Its characteristic fold has also been observed in other type I importers such as the E. coli maltose transporter [Citation25], methionine transporter [Citation26], and amino acid transporter [Citation27]. Based on crystal structures of type I ABC importers, it is known that the transmembrane domains possess at least two kinds of conformation involved in the substrate translocation cycle, an inward-facing conformation opening to the cytoplasmic space, and an outward-facing conformation opening to the periplasmic space. The strain A1 alginate transporter AlgM1M2SS is also homologous to type I ABC importers.
This review focuses on recent findings around alginate recognition and uptake in strain A1, in particular on the structural biology of the proteins involved in alginate incorporation.
Alginate recognition in strain A1
Alginate binding to cell surface protein p5
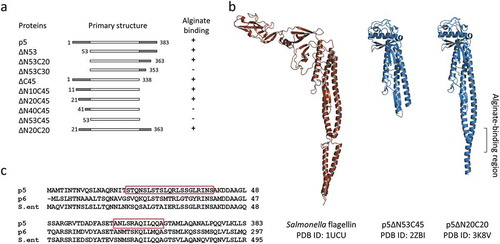
Strain A1 cell surface protein p5, homologous to a bacterial flagellin, binds tightly to alginate (Kd~10−9 M) at acidic pH [Citation14]. Because a similar interaction with alginate was also observed in E. coli flagellin, it is thought to be a universal property of flagellins. The p5 protein is able to entrap alginate at a locally acidic part of the cell surface, and thus is thought to play a role in concentrating alginate on the cell surface. Alternatively, the high affinity between p5 and alginate may imply that p5 is an alginate receptor. Flagellins are generally known for the diversity in their amino acid chain length [Citation28]. One of the most studied flagellins, FliC from Salmonella enterica, has about 500 amino acid residues, while p5 lacks about 100 amino acid residues in the center of the molecule.
The crystal structure of a p5 mutant lacking N-terminal 53 and C-terminal 45 amino acid residues (p5∆N53C45) (PDB ID: 2ZBI) consists of an α domain made up of three α helices and four β strands, and a β domain made up of eight β strands and two α helices [Citation29]. As predicted by primary sequence comparison, the structure of the α domain is similar to that of flagellins from other bacteria, while the structure of the β domain is quite different (). Three-dimensional structure and functional analyses of several deletion mutants of p5 suggested that amino acid residues 20–40 and 353–363 are essential for alginate binding. Amino acid sequences in these regions are highly conserved among bacterial flagellins, and contain several basic residues (). The lack of alginate binding by some deletion mutants is not due to structural disorders. In general, flagellins self-assemble in vitro; however, deletion of N-terminal and C-terminal amino acid residues from flagellin removes this ability. Because no correlation exists between self-assembly and alginate-binding ability, the polymerization of p5 is not essential for alginate recognition. The three-dimensional structure of S. enterica flagellin, as determined by cryo-electron microscopy, showed corresponding regions required for alginate binding in p5 form loop structures connecting two adjacent helices [Citation30]. We considered this small cleft formed between two α domains to be crucial for alginate binding. However, the crystal structure of p5 mutants that lacked N-terminal 20 and C-terminal 20 amino acid residues (p5∆N20C20) that retained alginate-binding ability but did not form filaments, showed there was no loop between the two helices but that they formed a continuous helix (unpublished result; PDB ID, 3K8V). The cleft structure formed between two α domains is unnecessary for alginate binding.
Figure 2. p5 and its derivatives.
(a) The correlation between amino acid deletions and alginate binding. (b) Tertiary structures of Salmonella flagellin and two p5 mutants. (c) Amino acid sequence alignment of N- and C-terminal regions of p5, p6, and Salmonella flagellin. Residues 20–40 and 353–363 of p5 are boxed.
Flagellin is a subunit protein composing bacterial flagellum for cell locomotion. Strain A1 was originally identified as a non-motile, non-flagellate bacteria [Citation31]. However, in the course of study on how p5 interacts on alginate on the cell surface, it has been found that strain A1 does have a flagellum-forming ability [Citation32,Citation33]. When non-motile wild-type strain A1 was subcultured on soft agar plates, motile cells, designated strain A1-M5, were observed [Citation32]. According to the position of flagella on a bacterial cell, they can be classified into several types, i.e., peritrichous, polar, sub-polar or lateral flagella [Citation34]. Strain A1-M5 cells possess a single uni-polar flagellum. Microarray analysis showed that mRNA of p5 in this strain increased 67-fold compared with the wild-type strain A1. In general, a bacterium possesses one flagellar system; however, some bacterial species e.g., Aeromonas, Azospirillum, Rhodospirillum and Vibrio, possess two distinct flagellar systems. Such bacteria constitutively form polar flagella expressed by one flagellar system and lateral flagella from the other system, depending on their growth conditions [Citation35,Citation36]. Some bacteria form one flagellum from multiple flagellins with high shared sequence homologies [Citation37]. On the other hand, flagellins with low shared sequence identities form different types of flagella, typically polar and lateral flagella [Citation35]. Through genome analysis of strain A1, it has been revealed that the bacterium possesses many flagella-related genes, including flagellin genes. The flagella-related genes in strain A1 can be divided into two gene sets [Citation32]. Gene set I is a large gene cluster containing 35 flagella-related genes, while gene set II contains small clusters of between two to twelve genes, and has a total of 46 flagellar genes [Citation32]. Two flagellins, p5 and p6, have been found in the outer membrane fraction of non-flagellated strain A1 grown on alginate medium [Citation14]. In addition to p5 and p6, another flagellin, p5ʹ, is also expressed at a lower level in strain A1-M5. As judged from their amino acid sequences, and as a result of promoter analyses, p6 is thought to be a lateral flagellin while p5 and p5ʹ are non-lateral flagellins. Although strain A1 possesses two distinct types (lateral and polar) of flagellar gene sets, the cells produce only a single polar flagellum. Strain A1 polar flagellar filaments consist of two kinds of polar flagellins (p5 and p5ʹ), as well as a lateral flagellin (p6), in contrast to the flagellar filaments of other bacteria. Interestingly, it was observed that flagellated strain A1 swarmed toward alginate on semi-solid agar plates, but not toward other polysaccharides [Citation33], indicating the existence of some type of alginate detection system in strain A1. As described above, p5 can interact closely with alginate. The possibility of a recognition mechanism for alginate via flagellar filaments is suggested.
Metal -binding protein Algp7
Another cell surface protein, Algp7, whose expression is induced in the presence of alginate, shows an affinity (Kd~10−8 M) specific to high molecular weight alginate [Citation38]. Algp7 shows homology to lipoprotein [Citation14] but lacks a lipobox [Citation38]. The gene coding for Algp7 along with three upstream genes forms an operon. This operon is homologous to that of a tripartite, low-pH ferrous ion transporter (EfeUOB) observed in E. coli [Citation39,Citation40]. The primary structures of EfeU, EfeO, and EfeB are similar to those of yeast Ftr1p iron permease, M75 metallopeptidase, and periplasmic peroxidase, respectively. Algp7 from strain A1 shows homology to EfeO, and has the iron binding HXXE motif.
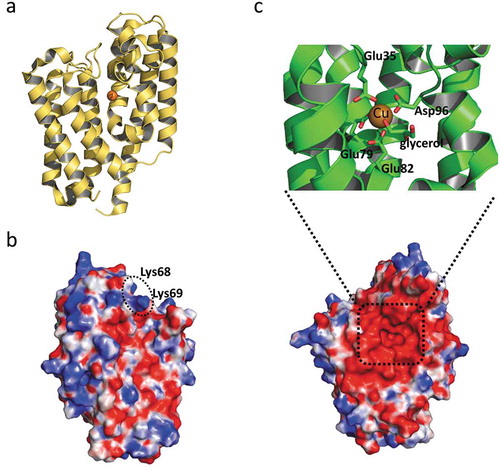
The three-dimensional structure of Algp7 (PDB ID, 3AT7, 5Y4C), solved by us, consists of two up-and-down helix bundles (). An ion network is formed at the metal binding motif and surrounds the molecular surface in a characteristic manner [Citation41]. Although Algp7 is categorized as an M75 metallopeptidase, no peptidase activity has been detected for this protein so far. Despite our efforts to crystallize Algp7 in complex with alginate, no crystal containing alginate has been obtained. Initially, the surface cleft with its ion network was considered to be an alginate-binding site [Citation41]. A mutational study and an in silico model of Algp7/oligoalginate constructed by a docking simulation using coordinates of Algp7 and alginate oligosaccharide suggested that two positively charged residues (Lys68 and Lys69) on another site of the molecule were necessary for alginate binding () [Citation42]. Algp7 also binds metal ions such as ferric, ferrous, and zinc ions. In the crystal structure (PDB ID, 5Y4C), an electron density map for a metal ion, probably copper, was observed on the opposite side to the proposed alginate-binding site. The metal ion was bound to the cleft between two bundles and coordinated by four acidic residues, i.e., Glu78, Glu82, Asp96, and Glu178 (), distinct from the M75 motif [Citation43]. Algp7 may deprive alginate of metal ions during the binding and release of the polysaccharide because alginate works as a chelator of dicationic ions.
Figure 3. The structure of Algp7 (PDB ID; 5Y4C).
(a) The overall structure of Algp7 with bound copper ion (orange ball). (b) and (c) The surface structure of Algp7. The proposed alginate-binding site is circled (b), the copper ion-binding site is shown in the inset (c). Blue and red in the surface model represent positively and negatively charged amino acid residues, respectively. The orientation of (c) is the same with that of (a), but opposite to that of (b).
Alginate uptake in strain A1
Heteropolysaccharide binding to periplasmic alginate-binding proteins
Solute-binding proteins essential for type I and II bacterial ABC importers are classified into several groups based on their three-dimensional structures [Citation44]. Alginate-binding proteins of strain A1, AlgQ1 and AlgQ2, are coded at the alginate uptake-gene operon and share about 85% of the same amino acid sequence with each other. They are larger in size compared with other solute-binding proteins. Among the more than 500 solute-binding proteins for which structures are known, a few proteins, including AlgQ1 and AlgQ2, have been categorized into the most recently identified seventh group. A member of this group, the FusA protein of Streptococcus pneumoniae, binds to fructo-oligo- and fructo-polysaccharides, such as kestose and inulin, and transfers them to an ABC importer [Citation45]. Members in this group may have the basic structure essential for interaction with polymers.
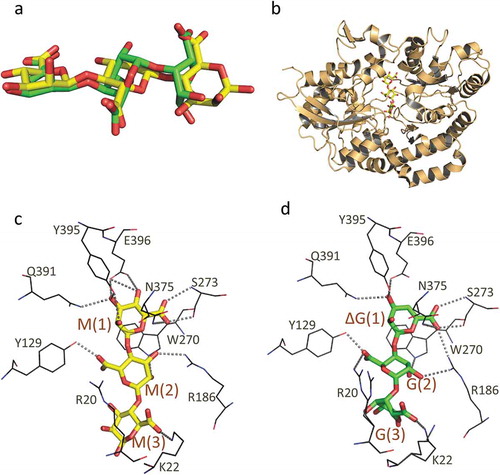
As described above, alginate is a linear acidic polysaccharide constituted by M and G, which are C-5 epimers of each other. Strain A1 cells can grow not only on alginate polysaccharide with different M/G ratios but also on oligoalginates. The M/G ratio does not influence the growth of strain A1. Alginate passed across the outer membrane is bound by the solute-binding proteins, AlgQ1 or AlgQ2, in the periplasm. AlgQ1 and AlgQ2 are thought to function in a similar way. Interactions between AlgQ1 or AlgQ2 and oligoalginates were observed by using a differential scanning fluorimetry method or ultraviolet absorption difference spectroscopy [Citation46,Citation47]. All alginate oligosaccharides examined interacted with both AlgQ1 and AlgQ2, regardless of differences in M/G composition, structure of the non-reducing end sugar (saturated or unsaturated), and sugar length (di- to tetrasaccharides). The dissociation constants (Kd) of some kinds of alginate oligomers were determined to be in the range of 10−6 ~ 10−5 M. Based on the crystal structures of AlgQ1 in complex with six types of oligoalginates (MMM, MG, ΔMMM, ΔGGG, ΔMMGM, and ΔMM) [Citation46,Citation47] (PDB ID; 3VLU, 3VLW, 3A09, 3VLV,1Y3P, 1Y3N) and AlgQ2 with ΔMMM and ΔMMGM [Citation48] (PDB ID; 5H71, 1J1N), it was found that two binding proteins with at least four subsites recognized the non-reducing end sugar residue of oligosaccharides most accurately, but did not prefer saturated G residues at subsite 1 because of steric hindrance of the C-5 carboxyl group. Although the structures of oligoalginates differ according to their composition, AlgQ1s with different oligoalginates are structurally identical even at their ligand-binding site (). AlgQ1 interacts flexibly with the hydroxyl groups of uronate residues at subsites 2 and 3, while C-5 carboxyl groups are recognized by specific residues of AlgQ1, although there is a difference in the orientation of the carboxyl group between G and M residues. This mechanism for substrate recognition enables AlgQ1 and AlgQ2 to bind to heteropolysaccharide alginate, leading to the sufficient growth of strain A1 cells in various alginates with different M/G ratios.
Figure 4. The structure of AlgQ1 and alginate oligosaccharide. (a) The structural difference between MMM and ΔGGG. Carbon atoms in MMM and ΔGGG are represented as yellow and green, respectively. (b) The overall structure of AlgQ1 in complex with MMM (PDB ID; 3VLU). (c) The alginate-binding site of AlgQ1 in complex with MMM (PDB ID; 3VLU). (d) The alginate-binding site of AlgQ1 in complex with ΔGGG (PDB ID; 3VLV). Hydrogen bonds formed between AlgQ1 and alginate are shown as dashed lines. The subsite number is shown in the parentheses in (c) and (d).
Alginate ABC importer
Alginates that are bound by AlgQ1 and AlgQ2 in the periplasm are transferred to the ABC importer. The alginate ABC importer of strain A1, AlgM1M2SS, comprises the transmembrane AlgM1-AlgM2 heterodimer and the cytoplasmic AlgS-AlgS homodimer. Genes coding for the three subunits AlgS, AlgM1, and AlgM2, together with other genes for related proteins such as periplasmic solute-binding proteins and alginate lyases, form a cluster in the strain A1 genome () [Citation49]. Three subunits, i.e., AlgS, AlgM1, and AlgM2, have been simultaneously expressed using the E. coli system and purified as a heterotetramer, using several detergents and a histidine tag connected to the C-terminus of AlgM2 [Citation50].
The importer transports alginate to the cytoplasm using ATP hydrolysis energy from the cytoplasmic AlgS. The ATP hydrolysis activity of AlgM1M2SS was increased by the addition of alginate or its oligosaccharide in the presence of an alginate-binding protein when AlgM1M2SS was reconstituted into liposomes or dissolved in a specific detergent solution [Citation50]. ATP hydrolysis activity of similar levels was detected using oligosaccharides with different M/G ratios, probably due to the wide substrate recognition ability of AlgQ2, as described above. On the other hand, cellotriose and chitotriose did not induce AlgM1M2SS activity [Citation50]. Furthermore, when fluorescently-labeled (2-pyridylaminated) alginate oligosaccharides (di- to tetrasaccharides) were added to proteoliposomes in the presence of AlgQ2, the transportation and ATP hydrolysis activity of AlgM1M2SS were increased [Citation50]. Vanadate, a phosphate analog, inhibited both ATP hydrolysis and substrate transportation. These observations indicate that purified AlgM1M2SS specifically transports alginate, depending on the presence of AlgQ1 or AlgQ2, and ATP hydrolysis by AlgS.
AlgM1M2SS tetramers purified using detergents were crystallized with and without an alginate-binding protein, nucleotide, and an oligoalginate. AlgM1M2SS tetramer, and AlgM1M2SS complexed with AlgQ2 and ΔMMM, were successfully crystallized and analyzed at resolutions of 4.5 Å and 3.2 Å, respectively [Citation50] (PDB ID; 4TQV, 4TQU). In the structural analysis, we used mutant proteins, a deletion mutant of AlgM1, in which N-terminal 24 residues were omitted, and an inactive mutant of AlgS where Glu160 had been replaced with Gln. The conformation of AlgM1M2SS in both crystals was essentially the same. AlgM1 and AlgM2 showed similar topology, with six transmembrane helices, and a dimer that is closed to the periplasm but open to the cytoplasm, the so-called inward-facing conformation (). One helix of AlgM2 protrudes into the periplasmic space and is therefore an important region for interactions with a solute-binding protein. Two helices in the cytoplasm are important for interactions between AlgS and AlgM1 or AlgM2. One of the two helices in each subunit fits with the pocket of AlgS. The helix is called the coupling helix and is conserved among ABC transporters. Two ATP-binding sites of the AlgSS homodimers are widely separated from each other, and the C-terminal regulatory domains interact with each other in the crystal structure. Considering these structural features, the conformation of AlgM1M2SS/AlgQ2 complex is believed to be in the state before substrate translocation. In the AlgM1M2SS/AlgQ2 complex, the C-terminal domain of AlgQ2 interacts with AlgM2, and the N-terminal domain interacts with AlgM1. The substrate-binding cleft between the two domains of AlgQ2 is partially covered by AlgM1M2. Therefore, a long tunnel-like structure is formed in the AlgM1M2SS/AlgQ2 complex (). The tunnel continues to the alginate-binding site of AlgQ2, and its length is about 30 Å, corresponding to alginate heptasaccharide. This tunnel-like structure is thought to be essential for the interaction between AlgM1M2SS and AlgQ2 binding alginate polymer.
Figure 5. The crystal structure of the alginate transporter (PDB ID; 4TQU). (a) The overall structure of AlgM1M2SS in complex with AlgQ2. (b) The tunnel structure (grayed mesh) that continues to the alginate-binding site of AlgQ2. (c) Charged amino acid residues in transmembrane AlgM1M2. Blue and red represent basic and acidic residues, respectively.
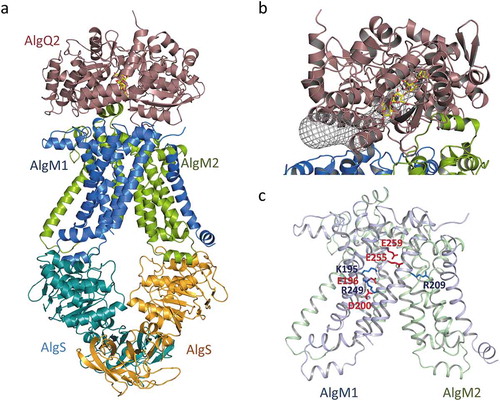
A large inner cavity in the transmembrane domain of the ABC transporter is used to translocate substrates across the cell membrane. This is also true for AlgM1M2SS. The surface of the inner cavity of AlgM1M2 contains both acidic and basic amino acid residues (); it is different from that of the maltose transporter which transfers neutral saccharides as a substrate. The importance of these charged residues for the effective binding of alginate from AlgQ1 or AlgQ2, and its subsequent release to the cytoplasm, was partially understood through mutagenesis analysis [Citation50]. In general, conformational changes of the ABC importer occur in accordance with the binding and release of solute-binding proteins and/or ATP hydrolysis of the nucleotide-binding domain, accompanied by the opening and closing motions of the periplasmic and cytoplasmic ends of the inner cavity, though the manner of conformational change is different for each type of ABC transporter. As a result of such conformational change, the ABC importer can carry substrate from the periplasm to the cytoplasm. In AlgM1M2SS, the distance between the periplasmic and cytoplasmic ends of the inner cavity is approximately 27 Å, corresponding to approximately six residues of linear alginate. The distinctive charged residues observed at the surface of the AlgM1M2 inner cavity are absent in MalFG, which transfers maltose. A site-directed mutagenesis study suggested that negative and positive charges derived from the residues Glu259 and Arg209 on the periplasmic side of AlgM1 and AlgM2 may contribute to the effective passing of the negatively charged substrate released by the alginate-binding protein. Other mutants at the surface of the inner cavity of AlgM1M2, such as H141A, K195A, E196A, and R249A of AlgM1, showed differing decreases in their transportation and ATP hydrolysis activities [Citation50]. These mutants might fail due to incorrect positioning of the substrate, resulting in inadequate conformational changes in AlgM1M2 upon ATP hydrolysis by AlgS.
In our assay system for alginate transportation activity, fluorescently-labeled oligoalginates taken into the proteoliposome containing AlgM1M2SS were quantified. Transportation of oligosaccharides longer than tetrasaccharides were not observed in this system. On the other hand, fluorescently-labeled longer oligosaccharides were able enhance the ATP hydrolysis activity of the proteoliposome. Since the crystal structure of the AlgM1M2SS/AlgQ2 pentamer in complex with longer oligoalginates adopts the same inward-facing conformation as that with trisaccharides, it is believed that substrate-bound AlgQ2 in a closed form docks with AlgM1M2SS to cause ATP hydrolysis of AlgS [Citation51].
The structures of the AlgM1M2SS and AlgM1M2SS/AlgQ2 complexes are similar to those of the maltose transporter in general; however, there are some key differences, as described above. In particular, the tunnel-like structure at the AlgQ2 and AlgM1M2 interface and the charged features of the inner cavity of the AlgM1M2 dimer are specific to the alginate ABC transporter, suggesting these structural features are essential for poly- and acidic saccharide transportation.
Concluding remarks
The characteristic mechanism to incorporate alginate has been applied to other fields. Modified strain A1 containing alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC) genes from Zymomonas mobilis produced 13.0 g/L ethanol in three days using alginate as the sole carbon source [Citation52], which was the first report on bioethanol production from alginate. Molecular transplantation of the strain A1 alginate incorporation system to dioxin-degrading Sphingomonas wittichii RW1 led to the formation of the cell surface pit and an increase in the dioxin-degrading capacity of the modified strain [Citation53]. Recent knowledge gained about the molecular mechanism of alginate recognition and uptake by strain A1 could further increase the capabilities and applications of strain A1.
No potential conflict of interest was reported by the authors.
Acknowledgments
Studies were performed with members of the Laboratory of Basic and Applied Molecular Biotechnology, Graduate School of Agriculture, Kyoto University.
Additional information
Funding
References
- Haug A, Larsen B, Smidsrød B. Uronic acid sequence in alginate from different sources. Carbohydr Res. 1974;32:217–225.
- Lee KY, Mooney DJ. Alginate: properties and biomedical applications. Prog Polym Sci. 2012;37:106–126.
- Szekalska M, Pucilowska A, Szymanska E, et al. Alginate: current use and future perspectives in pharmaceutical and biomedical applications. Int J Polm Sci. 2016;7697031:17.
- Rehm BH, Valla S. Bacterial alginates: biosynthesis and applications. Appl Microbiol Biotechnol. 1997;48:281–288.
- Boyd A, Chakrabarty AM. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J Ind Microbiol. 1995;15:162–168.
- Hay ID, Rehman ZU, Moradali MF, et al. Microbial alginate production, modification and its applications. Microb Biotechnol. 2013;6:637–650.
- Kawai S, Murata K. Biofuel production based on carbohydrate from both brown and red macroalgae: recent developments in key biotechnologies. Int J Mol Sci. 2016;17:145.
- Takagi T, Kuroda K, Ueda M. Platform construction of molecular breeding for utilization of brown macroalgae. J Biosci Bioeng. 2018;125:1–7.
- White DC, Sutton SD, Ringelberg DB. The genus Sphingomonas: physiology and ecology. Curr Opin Biotechnol. 1996;7:301–306.
- Kawasaki S, Moriguchi R, Sekiya K, et al. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J Bacteriol. 1994;176:284–290.
- Hisano T, Yonemoto Y, Yamashita T, et al. Direct uptake of alginate molecules through a pit on the bacterial cell surface: a novel mechanism for the uptake of macromolecules. J Ferment Bioeng. 1995;79:538–544.
- Hisano T, Kimura N, Hashimoto W, et al. Pit structure on bacterial cell surface. Biochem Biophys Res Commun. 1996;220:979–982.
- Hashimoto W, Momma K, Maruyama Y, et al. Structure and function of bacterial super-biosystem responsible for import and depolymerization of macromolecules. Biosci Biotechnol Biochem. 2005;69:673–692.
- Hashimoto W, He J, Wada Y, et al. Proteomics-based identification of outer-membrane proteins responsible for import of macromolecules in Sphingomonas sp. A1: alginate-binding flagellin on the cell surface. Biochemistry. 2005;44:13783–13794.
- Jones PM, George AM. The ABC transporter structure and mechanism: perspectives on recent research. Cell Mol Life Sci. 2004;61:682–699.
- Higgins CF. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113.
- Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Curr Opin Struct Biol. 2008;18:726–733.
- Locher KP. Structure and mechanism of ATP-binding cassette transporters. Philos Trans R Soc Lond B Biol Sci. 2009;364:239–245.
- Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. 2009;10:218–227.
- Oldham ML, Chen J. Snapshots of the maltose transporter during ATP hydrolysis. Proc Natl Acad Sci USA. 2011;108:15152–15156.
- Slotboom DJ. Structural and mechanistic insights into prokaryotic energy-coupling factor transporters. Nat Rev Microbiol. 2014;12:79–87.
- Locher KP. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol. 2016;23:487–493.
- Rice AJ, Park A, Pinkett HW. Diversity in ABC transporters: type I, II, and III importers. Crit Rev Biochem Mol Biol. 2014;49:426–437.
- Hollenstein K, Frei DC, Locher KP. Structure of an ABC transporter in complex with its binding protein. Nature. 2007;446:213–216.
- Oldham ML, Khare D, Quiocho FA, et al. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450:515–521.
- Kadaba NS, Kaiser JT, Johnson E, et al. The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science. 2008;321:250–253.
- Yu J, Ge J, Heuveling J, et al. Structural basis for substrate specificity of an amino acid ABC transporter. Proc Natl Acad Sci USA. 2015;112:5243–5248.
- Aizawa S. The flagellar world. NY: Academic Press; 2014.
- Maruyama Y, Momma M, Mikami B, et al. Crystal structure of a novel bacterial cell-surface flagel-lin binding to a polysaccharide. Biochemistry. 2008;47:1393–1402.
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650.
- Yonemoto Y, Murata K, Kimura A, et al. Bacterial alginate lyase: characterization of alginate lyase-producing bacteria and purification of the enzyme. J Ferment Bioeng. 1991;72:152–157.
- Maruyama Y, Kobayashi M, Murata K, et al. Formation of a single polar flagellum by two distinct flagellar gene sets in Sphingomonas sp. strain A1. Microbiology. 2015;161:1552–1560.
- Kobayashi M, Konishi H, Maruyama Y, et al. Lateral-typed flagellin responsible for formation of a polar flagellum but not of lateral flagella in Sphingomonas sp. strain A1. Microbiology. 2016;162:2042–2052.
- Leifson E. Atlas of bacterial flagellation. NY: Academic Press; 1960.
- McCater LL. Dual flagellar systems enable motility under different circumstances. J Mol Biol. 2004;7:18–29.
- Merino S, Shaw JG, Tomás JM. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiol Lett. 2006;263:127–135.
- Faulds-Pain A, Birchall C, Aldridge C, et al. Flagellin redundancy in Caulobacter crescentus and its implications for flagellar filament assembly. J Bacteriol. 2011;193:2695–2707.
- He J, Ochiai A, Fukuda Y, et al. A putative lipoprotein of Sphingomonas sp. strain A1 binds alginate rather than a lipid moiety. FEMS Microbiol Lett. 2008;288:221–226.
- Cao J, Woodhall MR, Alvarez J, et al. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157: H7. Mol Microbiol. 2007;65:857–875.
- Rajasekaran MB, Nilapwar S, Andrews SC, et al. EfeO-cupredoxins: major new members of the cupredoxin superfamily with roles in bacterial iron transport. Biometals. 2010;23:1–17.
- Maruyama Y, Ochiai A, Mikami B, et al. Crystal structure of bacterial cell-surface alginate-binding protein with an M75 peptidase motif. Biochem Biophys Res Commun. 2011;405:411–416.
- Temtrirath K, Murata K, Hashimoto W. Structural insights into alginate binding by bacterial cell-surface protein. Carbohydr Res. 2015;404:39–45.
- Temtrirath K, Okumura K, Maruyama Y, et al. Binding mode of metal ions to the bacterial iron import protein EfeO. Biochem Biophys Res Commun. 2017;493:1095–1101.
- Scheepers GH. Lycklama A Nijeiholt JA, Poolman B. An updated structural classification of substrate-binding proteins. FEBS Lett. 2016;590:4393–4401.
- Culurgioni S, Harris G, Singh AK, et al. Structural basis for regulation and specificity of fructooligosaccharide import in Streptococcus pneumoniae. Structure. 2017;25:79–93.
- Nishitani Y, Maruyama Y, Itoh T, et al. Recognition of heteropolysaccharide alginate by periplasmic solute-binding proteins of a bacterial ABC transporter. Biochemistry. 2012;51:3622–3633.
- Momma K, Mishima Y, Hashimoto W, et al. Direct evidence for Sphingomonas sp. A1 periplasmic proteins as macromolecule-binding proteins associated with the ABC transporter: molecular insights into alginate transport in the periplasm. Biochemistry. 2005;44:5053–5064.
- Mishima Y, Momma K, Hashimoto W, et al. Crystal structure of AlgQ2, a macromolecule (alginate)-binding protein of Sphingomonas sp. A1, complexed with an alginate tetrasaccharide at 1.6-Å resolution. J Biol Chem. 2003;278:6552–6559.
- Momma K, Okamoto M, Mishima Y, et al. A novel bacterial ATP-binding cassette transporter system that allows uptake of macromolecules. J Bacteriol. 2000;182:3998–4004.
- Maruyama Y, Itoh T, Kaneko A, et al. Structure of a bacterial ABC transporter involved in the import of an acidic polysaccharide alginate. Structure. 2015;23:1643–1654.
- Kaneko A, Uenishi K, Maruyama Y, et al. A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate. J Biol Chem. 2017;292:15681–15690.
- Takeda T, Yoneyama F, Kawai S, et al. Bioethanol production from marine biomass alginate by metabolically engineered bacteria. Energy Environ Sci. 2011;4:2575–2581.
- Aso Y, Miyamoto Y, Harada KM, et al. Engineered membrane super-channel improves bioremediation potential of dioxin-degrading bacteria. Nat Biotechnol. 2006;24:188–189.

