Abstract
At present, the planet faces a change in the composition and bioavailability of nutrients. Zinc deficiency is a widespread problem throughout the world. It is imperative to understand the mechanisms that organisms use to adapt to the deficiency of this micronutrient. In the Ascomycetes fungi, the ZIP family of proteins is one of the most important for zinc transport and includes high affinity Zrt1p and low zinc affinity Zrt2p transporters. After identification and characterization of ZRT1/ZRT2-like genes in Ustilago maydis we conclude that they encode for high and low zinc affinity transporters, with no apparent iron transport activity. These conclusions were supported by the gene deletion in Ustilago and the functional characterization of ZRT1/ZRT2-like genes by measuring the intracellular zinc content over a range of zinc availability. The functional complementation of the S. cerevisiae ZRT1Δ ZRT2Δ mutant with U. maydis genes supports this as well. U. maydis ZRT2 gene, was found to be regulated by pH through Rim101 pathway, thus providing novel insights into how this Basidiomycota fungus can adapt to different levels of Zn availability.
Introduction
Zn deficiency is a serious problem worldwide (Alloway, Citation2008; Hamid & Ahmad, Citation2001; Kabala & Singh, Citation2001; Velu et al., Citation2011). Poor food supply and cereal-based diets with low micronutrient content, are considered to be major causes of nutritional deficiencies affecting 3 billion humans and contributing to over 450,000 children deaths each year (Biesalski, Citation2013; Cakmak et al., Citation2010; Eide, Citation2006; Hotz & Brown, Citation2004; Kramer & Clemens, Citation2005; Stein et al., Citation2007; Welch & Graham, Citation2004). Zinc bioavailability for plants depends on many factors like Zn content in soil, pH, organic matter, soil temperature and moisture regimes, root distribution, and rhizosphere effects including microbial composition (Adiloglu & Adiloglu, Citation2006; Alloway, Citation2004, Citation2008, Citation2009).
An alternative mechanism to improve zinc bioavailability for plants is through their association with mycorrhizas, which increase the surface area for nutrient uptake (Baslam et al., Citation2014; Cavagnaro, Citation2008; Raklami et al., Citation2019). Several fungi belonging to the Basidiomycota division, form symbiotic associations with plants (Jin et al., Citation2019), and most of the mechanisms used by these fungi to obtain nutrients have not yet been described. U. maydis is a Basidiomycete fungus that causes the common disease known as corn smut (Ruiz-Herrera, Citation2008), and it is also used as a fungal model system to study how other organisms face challenging environmental conditions such as variations in temperature, humidity, pH, and nutrient availability, including metals like zinc (Kämper et al., Citation2006).
Zinc is an essential micronutrient for all organisms because it plays key roles as a structural or catalytic cofactor of numerous proteins within several cellular processes including replication, transcription, RNA processing, metabolism, and cell differentiation (Tapiero & Tew, Citation2003; Kramer & Clemens, Citation2005; Figueiredo et al., Citation2012). Organisms can be adversely affected not only by zinc-limiting conditions, but also by Zn excess. The optimum zinc content for cell growth is called the “zinc quota”, and in yeast, it has been estimated to be approximately 107 atoms per cell (Eide, Citation2006). In the last few years, it has been found that cells achieve intracellular zinc homeostasis by fine-tuning metal transport systems (Amich et al., Citation2014; MacDiarmid et al., Citation2000; Zhao & Eide, Citation1996a, Citation1996b).
An important group of zinc and/or iron transporters is the ZIP superfamily. ZIP transporters are found at all phylogenetic levels including bacteria, fungi, plants, and mammals (Gaither & Eide, Citation2001). Two of the earliest known ZIP representatives are transporters located at the plasma membrane of S. cerevisiae, Zrt1p and Zrt2p. Zrt1p has a high affinity for zinc, while Zrt2p has a lower affinity (Zhao & Eide, Citation1996a, Citation1996b). Both are expressed in metal-limiting conditions. Their activity and regulation have been described in Ascomycetes fungi, such as S. cerevisiae. In zinc-limiting medium at acidic pH their expression is induced by the transcription factor Zap1p; however, this induction does not occur in alkaline pH (Lamb et al., Citation2001; Lamb & Mitchell, Citation2003; Serrano et al., Citation2002). In Aspergillus nidulans, and in contrast to S. cerevisiae, the transcription of the orthologous genes ZRFA and ZRFB, is reduced at neutral or alkaline zinc-limiting medium by the PacCp transcription factor (Amich et al., Citation2009). Characterization of orthologue proteins in organisms of the Basidiomycota phylum is scarce due to their complexity in life cycles and growth at lab conditions (de Mattos-Shipley et al., Citation2016).
Here, we report the identification and characterization of two zinc transporters from U. maydis expressed during zinc starvation conditions. UmZrt1p (GenBank accession number MF177953) behaves as a high affinity transporter, while UmZrt2p (GenBank accession number MF177954) shows properties of a lower affinity transport for zinc. In our conditions, both proteins showed no ability to transport iron. Furthermore, we demonstrate that the transcription factor Rim101/PacCp is required for the repression of UmZRT2 in acidic environments but acts as an activator in neutral pH. UmZRT1 on the other hand, is not PacC/Rim101p-responsive. Our results provide novel insights into how U. maydis achieves zinc homeostasis and offer new avenues to improve zinc nutrition outside of traditional fungal model systems such as budding yeast.
Materials and methods
Strains of U. maydis used were wild-type FB1 (a1b1) (Banuett & Herskowitz, Citation1989), BMA2 (a2b2ΔRIM101::hyg) (Aréchiga & Ruiz-Herrera, Citation2005) ΔUmZRT1 (a1b1ΔZRT1::hyg) and ΔUmZRT2 (a1b1ΔZRT2::hyg). From S. cerevisiae, BY4743 (MATα/a his3/his3 leu2/leu2 ura3/ura3met15/MET15 lys2/LYS2) (Brachmann et al., Citation1998) and ZHY3 (MATα ade6 can1 his3 leu2 trp1 ura3 zrtl::LEU2 zrt2::HIS3) (Zhao & Eide, Citation1996a) were used. Escherichia coli strain was DH5α (F– Φ80lacZΔM15Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1) (Invitrogen, Carlsband, CA).
S. cerevisiae cells were grown in standard culture media (yeast minimal media/synthetic defined (SD) and yeast extract peptone dextrose (YPD)) supplemented with necessary auxotrophic requirements and 2% glucose (Amberg et al., Citation2005). U. maydis cells were grown in complete medium (Holliday, Citation1961) and YPD; both fungi were subjected to zinc-limiting conditions (Low zinc medium, LZM) using different concentrations of ZnCl2 (Eide & Guarente, Citation1992; Zhao & Eide, Citation1996a). E. coli was grown in Luria Bertani medium (Sezonov et al., Citation2007). Cell number in liquid cultures was determined measuring the optical density at 600 nm (OD600) in a Beckman DU800 spectrophotometer and using a Multiskan MCC 355 microplate reader (Thermo Fisher Scientific, Waltham, MA).
Zinc transport activity assessment by functional complementation in S. cerevisiae
DNA fragments of the UmZRTI and UmZRT2 open reading frames were amplified by PCR, from the FB1 strain using primers with flanking sequences for recombination with pDR195 plasmid. Oligonucleotides used are shown in Supplementary data (primers 1 and 2, Table S1). Resulting fragments were inserted into Not1-linearized pDR195 plasmid by recombination in vivo in S. cerevisiae BY4743. pDR195 contains the strong PMA1 promoter, ampicillin-resistance, and URA3 selection markers (Rentsch et al., Citation1995). Plasmids were then transformed into E. coli DH5α by standard methods (Green & Sambrook, Citation2012). Constructions were confirmed by restriction analysis (SalI and EcoRV enzymes for UmZRT1 and NdeI and EcoRI for UmZRT2) and by DNA sequencing using the primers 3 and 4 for UmZRT1 and 3, 4, 5 for UmZRT2 (Table S1).
The S. cerevisiae ZHY3 strain was transformed with the UmZRT1 and UmZRT2 constructions. pMC5 plasmid containing a S. cerevisiae genomic ZRT1 fragment was used as positive control and the pDR195 empty vector served as negative control. The ability of UmZRT1 and UmZRT2 to confer zinc uptake activity in ZHY3 was tested. The experiment was carried out in two different biological experiments, with triplicate cultures in each treatment. Cells were grown first in zinc-replete Synthetic Defined medium (SD) (Amberg et al., Citation2005) at 30 °C, and then washed and inoculated (starting at OD600 = 0.01) into liquid LZM supplemented with different ZnCl2 concentrations (1, 10, 100 and 1000 µM) and grown for 15 h. OD600 was then measured for each culture and results were validated through descriptive and inferential analysis, with the use of the program SPSS version 17 (SPSS Inc, Citation2008). The ANOVA and Tukey tests were performed to determine significant differences in absorbance mean values. Growth of these cells was also assessed by drop assay; 5 µl drops containing 1 × 105, 1 × 104, 1 × 103 and 1 × 102 cells of each transformant were applied on to solid LZM with concentrations mentioned before (Eide & Guarente, Citation1992; Zhao & Eide, Citation1996a).
Generation of Ustilago maydis mutants
Mutants of ZRT1 and ZRT2 genes in Ustilago maydis (FB1) were obtained using the split marker technique (Catlett et al., Citation2003; Fairhead et al., Citation1996, Citation1998). The generation of the gene replacement cassettes was performed as described previously by (Yu et al., Citation2004). As shown in Figure S1, using the primers listed in Table S2, the cassettes were constructed by homologous recombination of the region upstream and downstream of the gene of interest ORF and hygromycin B phosphotransferase gene (hph), afterwards using the cassette as template we amplified two fragments: from the start of the 5´ region to the middle of the selective marker and then from the middle of selective marker to the final of the 3´ region, transforming Ustilago maydis protoplast with these two fragments to get a triple recombination and less background during transformant selection. Mutations were confirmed by PCR, enzyme restriction analysis, and sequencing PCR fragments from the genome region.
Functional characterization in Ustilago maydis
For gene functional determination, U. maydis wild-type (a1b1), ΔUmZRT1 and ΔUmZRT2 strains were subjected to nutrient stress; first they were grown in YPD medium for 24 h, then washed twice with deionized water. Cells were resuspended in 5 ml of deionized water and incubated for 15 h, and then washed again twice with deionized water before their inoculation. The treatments were YNB supplemented with 0, 1, 10, 100 and 1000 µM of ZnCl2, and three different biological experiments were performed, starting the inoculation at OD600 = 0.5 and incubated for 15 h at 28 °C, subsequently washed three times with 5 mM ethylenediaminetetraacetic acid (EDTA)-25 mM Tris (pH = 8), and three times with deionized water to eliminate extracellular metals. The cell pellets were dried for 3 days at 60 °C and dry weights were determined. Digestion was accomplished with HNO3 trace metal grade (Fisher Scientific, Hampton, NH) and we proceeded with micronutrient (Fe, Mn, Zn) quantification by inductively coupled plasma-optical emission spectroscopy (ICP-OES) (protocol modified from Mendoza-Cózatl et al., Citation2014). Data were validated through one way ANOVA followed by Tukey multiple comparisons test using GraphPad Prism Version 6.01 (La Jolla, CA).
Ustilago maydis cellular drop assay
To characterize qualitative zinc transport capability, cellular drop assay was made in YNB with 0, 1, 10, 100, and 1000 µM of ZnCl2 added. The inoculated cells were subjected to the same nutritive stress than the analyzed by ICP-OES, and also started at OD600 = 0.5 and then serial dilutions were inoculated.
RNA isolation and gene expression analysis
U. maydis FB1 (WT) (Banuett & Herskowitz, Citation1989) and BMA2 (RIM101-) (Aréchiga & Ruiz-Herrera, Citation2005) strains were cultured first in SD medium overnight (12 h) at 28 °C, washed and then inoculated (starting at OD600 = 0.01) into LZM medium modified to expose cells to the same free zinc concentration at the pH values provided. The acidic medium was at pH 4.0 (LZM4) and contained 1 mM EDTA as a zinc-buffering chelator. The pH 7.0 medium (LZM7) was prepared with 1 mM ethyleneglycoltetraacetic acid (EGTA) as the chelator. The free zinc concentration was estimated to be ∼1 x10−12 M in both media using the MaxChelator program (Bers et al., Citation2010; Zhao & Eide, Citation1996a, Citation1996b). Following 15 h growth, total RNA was obtained with standard methods (Green & Sambrook, Citation2012) and treated with RQ1 RNase free DNase (Promega, Madison, WI) to eliminate any trace of DNA following the manufacturer protocol. For the cDNA synthesis, random hexamers were used (QIAGEN, Hilden, Germany). Real-Time Quantitative PCR (qPCR) assay was implemented to analyze the gene expression of Actin, UmZRT1, UmZRT2, NRG1 and RIM101 genes of FB1 and BMA2 strains, in two biological experiments and triplicates of each gene, using SsoAdvanced™ Universal SYBR® Green Supermix (BIO-RAD, Hercules, CA) in a CFX96 Touch™ Real Time PCR Detection System (BIO-RAD, Hercules, CA). The primer sets used are shown in Table S3 and all the reactions for the qRT-PCR were carried out as described in the manufacturer protocol of the reagents mentioned before. Relative gene expression quantification was performed with the Livak method (Livak & Schmittgen, Citation2001) using FB1 as calibrator and actin as reference gene for normalization.
Proteins structure prediction
For the prediction of proteins topology, we used the University College London PSIPRED server with the MEMSAT-SVM algorithm (Buchan et al., Citation2013), and then we compared the results with the transmembrane domain prediction programs ExpasyTMpred (Hofmann & Stoffel, Citation1993), HMMTOP (Tusnády & Simon, Citation2001), Protter version 1.0 (Omasits et al., Citation2014) and finally with the predicted structure annotated in UniProt database (The UniProt Consortium, Citation2018). In order to predict the protein cellular location, we use the DeepLoc1.0: Eukaryotic protein subcellular localization predictor (Almagro Armenteros et al., Citation2017). The prediction of functional domains and determination of identity percentages was carried out through alignments using the NCBÍs Basic Local Alignment Search Tool (BLAST®) and protein BLAST algorithm (NCBI Resource Coordinators, Citation2016).
Prediction of transcription factor binding in UmZRT1 and UmZRT2 promoters
In order to determine if Rim101p and Nrg1p influence the expression of the interest genes, 1000 bp upstream of the UmZRT1 and UmZRT2 ORFs were analyzed in the YEASTRACT database (Yeast Search for Transcriptional Regulators and Consensus Tracking) (Teixeira et al., Citation2018).
Results
Identification and analysis of genes that encode zinc transporters in U. maydis
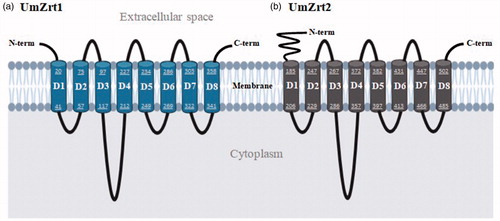
In silico analysis revealed two genes with putative zinc transport function in the U. maydis genome sequence (Kämper et al., Citation2006; Mewes et al., Citation2004). These genes correspond to the open reading frame identification numbers um00096 and um03110, which based on their predicted function were re-named to UmZRT1 and UmZRT2, respectively. These genes are located on chromosome 1 (UmZRT1) and 7 (UmZRT2) and their predicted topology of both proteins, UmZrt1p and UmZrt2p, suggested eight putative transmembrane domains on the plasma membrane, with a large cytosolic loop between the 3rd and 4th domains. Both of their amino and carboxyl termini are predicted to face the extracellular space. UmZrt2p displayed a larger extracellular amino acid chain before the first transmembrane domain compared with umZrt1p (. Structure predictions were performed as described in Materials and Methods and showed slight variations between prediction methods regarding the endpoints of amino acids in each transmembrane domain (Table S4).
Figure 1. In silico prediction of UmZrt1p and UmZrt2p membrane topology. Both proteins a) UmZrt1 and b) UmZrt2 have eight predicted transmembrane domains (D1-D8) with the N- and C-terminal ends facing the extracellular space. The numbers shown in each domain correspond to the amino acids that make them up.
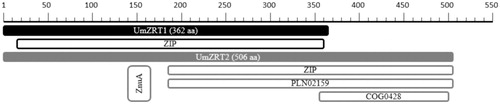
A comparative analysis of the U. maydis putative zinc transporters showed expected high identity percentage with other metal transporters. For instance, UmZRT1 coding sequence (GenBank accession number MF177953) predicts a protein of 362 amino acids and most of UmZrt1p’s amino acid sequence (amino acids 18-359) has 32.81% of identity percentage to the characteristic functional domain of the ZIP family. Similarly, the CDS of the UmZRT2 gene (GenBank accession number MF177954) predicts a 506 amino acid protein and NCBI Basic Local Alignment Search Tool (BLAST) identified the presence of various conserved domains. One domain corresponds to the typical ZIP superfamily (aa 182-506) with 32.54% of identity, a second domain (aa 182-506) has 29.79% of identity to PLN02159 in Arabidopsis, characteristic of Fe2+ transporter proteins and a third domain (aa 352-502) is related to COG0428 of the cl00437 superfamily with 29.50% of identity, whose hypothetical function is divalent metal transport in Archaea organisms. Finally, amino acids 141-166 of UmZrt2p also share 36% identity with a portion of the periplasmic ZnuA component of high affinity ZnuABC zinc transporters found in prokaryotic organisms (NCBI Resource Coordinators, Citation2016) (Finn et al., Citation2014) (.
UmZrt1p and UmZrt2p amino acid sequences share similarity with zrt/irt proteins from other organisms
By amino acid sequence comparison analysis through the NCBI’s BLAST tool and blastp algorithm, we found that proteins from various other organisms have significant percentages of identity with UmZrt1p and UmZrt2p. However, major differences were also found. For instance, at the amino acid level, these Ustilago maydis proteins showed only 30–44% of identity compared to Zrt proteins from S. cerevisiae as is shown in the amino acid sequences alignments (Figure S2, S3). Furthermore variations with Arabidopsis thaliana and Mus musculus were found despite having the typical ZIP functional domain (. Also, UmZrt1p only shared 31% of identity with its putative homolog UmZrt2p. These major differences prompted us to functionally characterize UmZrt1p and UmZrt2p at the molecular and physiological level.
Functional characterization of UmZRT1 and UmZRT2 by complementation assay in S. cerevisiae
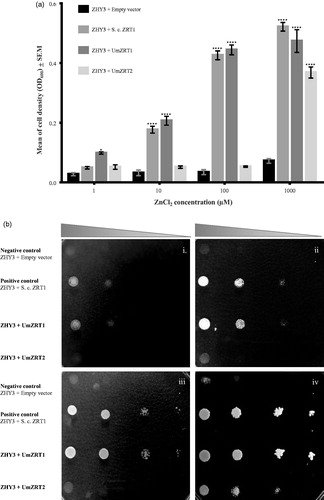
To determine whether the functions of the UmZRT1 and UmZRT2 genes are equivalent to those described for ZRT1 and ZRT2 in S. cerevisiae, the open reading frames of these U. maydis genes were amplified and cloned into the shuttle plasmid pDR195 suitable for expression in S. cerevisiae and driven by the strong PMA1 promoter (Rentsch et al., Citation1995). After sequence confirmation, these plasmids were used to test for functional complementation in the double mutant S. cerevisiae strain ZHY3 (zrt1Δ zrt2Δ). Growth analysis in liquid low zinc media (LZM) was performed over a range of zinc concentrations. UmZRT1 and UmZRT2 were transformed into ZHY3 and their growth was independently measured in triplicate experiments for each set of transformants. shows the means of cell densities of two independent transformations per construct after 15 h of culture. While the ZHY3 cells transformed with empty vector grew poorly at all tested zinc concentrations, cells expressing UmZRT1 showed a dose response growth similar to S. cerevisiae ZRT1-expressing cells. In contrast, cells complemented with UmZRT2 showed no complementation phenotype at low zinc concentrations (1–100 µM), but strong complementation at a higher concentration of ZnCl2 (1000 µM) (. Similar zinc dose dependence was observed for complementation on this mutant strain with S. cerevisiae ZRT2 (Zhao & Eide, Citation1996b). These results suggest that both UmZRT1 and UmZRT2 encode functional zinc transporters and provide evidence suggesting that UmZrt1p is a high affinity zinc transporter, similar to S. cerevisiae Zrt1p, while UmZrt2p has a lower affinity for zinc. These assays were also performed on solid media (LZM) and results were consistent with those obtained from liquid cultures (). While S. cerevisiae ZRT1 and UmZRT1 complemented the mutant strain at 1, 10, 100 and 1000 µM added zinc, UmZRT2 only presented complementation when 1000 µM of zinc was added.
Figure 4. Complementation assays of ability to transport zinc in Saccharomyces cerevisiae cells expressing Ustilago maydis zinc transporters. a) ZHY3 (zrt1Δ zrt2Δ) cells were transformed with the pDR195 vector (negative control), pMC5 (positive control, S. cerevisiae ZRT1), pDR195-UmZRT1, and pDR195-UmZRT2. Each bar represents the mean of the triplicates of two biological experiments for each treatment and the statistical difference between treatments and the negative control is shown (*p < 0.05 and ****p < 0.0001). b) Yeast transformants described before (a) were inoculated onto LZM plates in drops containing 1 × 105, 1 × 104, 1 × 103, and 1 × 102 cells. Panels i, ii, iii and iv correspond to LZM plus 1, 10, 100, 1000 µM ZnCl2, respectively.
Functional characterization of UmZRT1 and UmZRT2 genes in Ustilago maydis
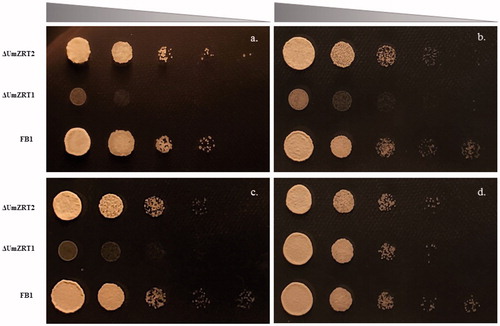
To gain more insight into the native function of UmZRT1 and UmZRT2, we generated mutants where the corresponding ORF in U. maydis was replaced by homologous recombination of a selection marker. After confirming by PCR and sequencing that the insertions had taken place at the correct loci, we performed elemental composition of cells by ICP-OES. shows that ΔUmZRT1 cells grown in YNB for 15 hrs without additional zinc added presented levels of Zn below the limit of detection (0.005 ppm) and significantly different compared to the wild-type strain FB1 (). Interestingly, ΔUmZRT2 have detectable levels of Zn but these were also significantly lower than FB1. A similar pattern was observed with cells grown in YNB with additional 1, 10 and 100 µM of ZnCl2 (Figure S4). In YNB media containing additional 1000 µM of ZnCl2, ΔUmZRT1 showed a slight increase of zinc concentration, compared to YNB without Zn added, while ΔUmZRT2 showed higher zinc concentrations although these were still significantly lower than wild-type (). Notably, deletion of either UmZRT1 or UmZRT2 had no impact on iron accumulation compared to wild-type (Figure S5), suggesting that these proteins play little or no role in the transport of iron. Manganese levels were under the limit of detection for all strains. Altogether, these results support the hypothesis that UmZRT1 encodes a high zinc affinity transporter while UmZRT2 a low zinc affinity transporter as predicted by sequence homology and functional complementation experiments in S. cerevisiae.
Figure 5. Quantification of zinc by ICP-OES. a) Determination of zinc abundance in dry weight of cells grown in YNB without zinc addition. b) Measure of zinc content in dry weight of cells grown in YNB supplemented with 1000 µM ZnCl2. Each bar represents the mean of the biological triplicates in each treatment and the statistical difference between FB1 and ZRT mutants is shown (**p < 0.01 and ***p < 0.001).
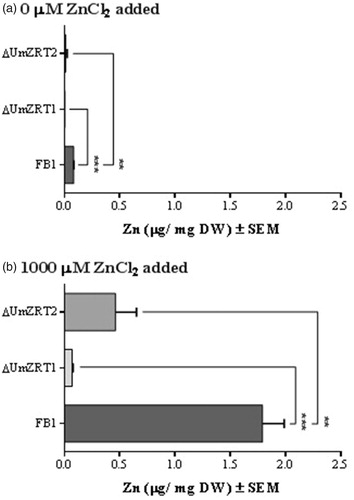
The growth ability of U. maydis wild-type and UmZrt1 and UmZrt2 mutants was also determined on solid YNB plates supplemented with 1, 10, 100, and 1000 µM of ZnCl2. ΔUmZRT1 showed less growth than the ΔUmZRT2 or the wild-type strain except at the highest zinc concentration tested where ΔUmZRT1 showed growth comparable to the other strains. These results were consistent with a defective high affinity zinc transporter, and further supports that UmZRT2 encode a low affinity zinc transporter (.
In silico analysis of the UmZRT1 and UmZRT2 promoters
Because the regulation of ZRT-like genes by Zap1p and PacC/Rim101p transcription factors has been reported previously, through in silico analysis, it was possible to look for potential binding sites for these transcription factors in the regions upstream of the UmZRT1 and UmZRT2 open reading frames. The UmZRT1 promoter region did not have any consensus Rim101/PacCp binding sites but did have several putative Nrg1p ones. Nrg1p is known to be regulated by Rim101p in S. cerevisiae (Vyas et al., Citation2005). In contrast, the UmZRT2 promoter had possible binding sites for both Rim101p and Nrg1p transcription factors. It should be noted that unlike S. cerevisiae ZRT1 and ZRT2 promoters, U. maydis genes do not contain any of the previously reported consensus sequences for Zap1p transcription factor binding; which is responsible for regulating many of the zinc transporters genes in most fungal organisms studied to date (Zhao et al., Citation1998) ().
Table 1. Location of potential binding sites for transcription factors with possible regulatory functions in the U. maydis zinc transporter UmZRT1 and UmZRT2 promoter regions. Consensus binding sites for Nrg1p and Rim101p transcription factors in UmZRT1 and UmZRT2 promoters and their distance (bp) upstream relative of the ATG initiation codon.
UmZRT1 and UmZRT2 expression analysis
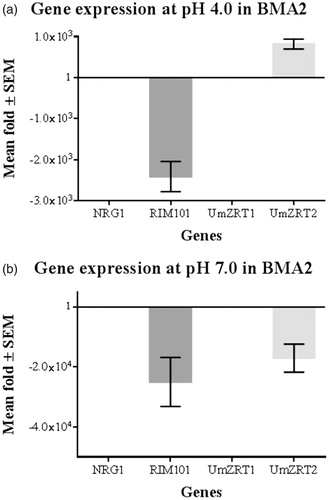
To evaluate UmZRT1 and UmZRT2 relative expression levels in response to changes in zinc concentration under acidic and neutral conditions, we measured their mRNA levels along with those of RIM101 and NRG1 in a wild-type (FB1) and BMA2 strain (ΔRIM101). In order to obtain the fold change in expression of these genes, data were analyzed by Livak method. As described in Materials and Methods actin gene was used as reference gene, and constant levels of its expression were exhibit in all samples. The calibrator used was FB1, and after normalization the fold change of the control was 1 as a constant. Finally data of the controls were incorporated in the interest genes fold change. At acidic pH (LZM pH 4.0) UmZRT2 was increased in expression ∼800-fold in BMA2 compared with the FB1 calibrator, which suggests that PacC/Rim101p is responsible of UmZRT2 repression at low zinc concentration and acidic pH in the wild-type strain (FB1). UmZRT1 and NRG1 did not show significant expression variations at this pH (). Under neutral conditions (LZM pH 7.0), no expression differences were observed of the measured genes except UmZRT2, which presented repression of greater than 1000-fold compared with calibrator. This result, revealed the potential role of Rim101p as activator of UmZRT2 expression also at neutral pH ().
Figure 7. Relative gene expression of BMA2 at acidic and neutral pH. Each bar represents the mean fold change (by Livak method) for triplicate measurements of two biological experiments corresponding to each gene in FB1 (calibrator) and BMA2 (ΔRIM101). The fold change of the control is 1 as a constant and data of the control are incorporated in the interest genes fold change. a) Fold differences in expression at pH 4.0 and low zinc concentrations in BMA2 compared with FB1. Repression effect of Rim101p on UmZRT2 is shown. b) Fold differences in expression at pH 7.0 and low zinc concentrations of BMA2. The activator effect of Rim101p on FB1 is exhibited, since BMA2 shows more than 1000 fold of repression at this condition.
Discussion
Intracellular ion homeostasis is essential for a stable physiology of living cells, so organisms have evolved different strategies to achieve such homeostasis. One strategy often used by organisms across different phyla is the expression of transporters with different substrate affinities to traffic them into the cytosol, organelles, or to the extracellular space. The ZIP family of transporters (Zrt/Irt-like Proteins) was first identified in plants and their members transport various metals, such as cadmium, iron, manganese, and/or zinc (Guerinot, Citation2000), from the extracellular space or organellar lumen into the cytosol (Balaji & Colvin, Citation2005). ZIP transporters have been described in organisms belonging to diverse phyla (Eide, Citation2005; Gaither & Eide, Citation2001). Founding members of the ZIP family include the S. cerevisiae Zrt1p and Zrt2p (zinc-regulated transporters) proteins, which have been well characterized (Zhao & Eide, Citation1996a, Citation1996b). Also, ZIP orthologs are reported in other fungi, mainly in other Ascomycetes, such as A. fumigatus (ZrfA and ZrfB) (Amich, Citation2010). However, the study of ZIP orthologs in Basidiomycetes remains scarce (Wilson et al., Citation2012).
In this work, two Zn2+ uptake systems in the Basidiomycete U. maydis, corresponding to accession number MF177953 (UmZRT1) and MF177954 (UmZRT2), were identified by amino acid similarity with other transporters reported. Similarity with members of the ZIP family was evident by the presence of ZIP domains in both proteins (. Most ZIP transporters have similar topologies, characterized by the presence of eight predicted transmembrane domains, with N- and C-termini located on the outside of the plasma membrane (Nishida et al., Citation2008). ZIP proteins can range widely in length; main differences in length are located between domains III and IV (termed the “variable region”), which has a potential metal-binding site located in the cytoplasm, and at their N-terminal ends (Guerinot, Citation2000). In U. maydis, we found by in silico analysis that these proteins likely contain eight transmembrane domains, located in the plasma membrane with N- and C terminal chains on the extracellular side and both with an extensive variable region between the domains mentioned before, on the cytosolic side and marked differences in length and amino acid content at their N-termini (. UmZRT1 encodes a protein of 362 amino acids and UmZRT2 consists of 506 amino acid residues. Length differences are primarily due to the mentioned N-termini chains variations, which are 19 residues long for UmZrt1p and 184 residues long for UmZrt2. When compared with S. cerevisiae Zrt1/Zrt2, these proteins have more identity and similarity at the ZIP domain (Figure S2 and S3) (Buchan et al., Citation2013; The UniProt Consortium, Citation2018).
Kinetic studies of zinc uptake by S. cerevisiae cells grown with different amounts of substrate in the medium had suggested the distinctive functionalities of the two main zinc transporters; Zrt1p has a high affinity for Zn2+ and is only active in zinc limited cells (Zhao & Eide, Citation1996a) and Zrt2p has a lower affinity system for zinc uptake and is more active in zinc-replete cells (Zhao & Eide, Citation1996b). Our complementation analysis suggested that UmZRT1 gene is an ortholog of S. cerevisiae ZRT1, as it was able to restore metal transport in the ZHY3 (ZRT1Δ ZRT2Δ) strain at low ZnCl2 concentrations (. Moreover, growth of the ΔUmZRT1 strain was severely affected at low Zn concentrations (). Additionally, we found that UmZRT2 gene encodes a transporter whose affinity for Zn2+ appears to be lower than UmZRT1, because complementation was only observed at high zinc concentrations (1000 μM ZnCl2) (), and growth of the ΔUmZRT2 was less affected at different Zn concentrations tested (. Thus, UmZrt2 is a likely ortholog of the low affinity S. cerevisiae Zrt2 transporter. The intracellular zinc content measured after growth with increasing ZnCl2 concentrations in Ustilago maydis wild-type, ΔUmZRT1, and ΔUmZRT2 strains by ICP-OES further supported this hypothesis of high and low zinc affinities for UmZRT1 and UmZRT2, respectively. Notably, there were no variations observed in the content of another metal commonly transported by ZIP members (i.e. iron), so these proteins are not likely to be involved in iron transport (Figure S5).
In all organisms, zinc uptake is controlled to ensure the optimum levels of this metal, while preventing its potentially toxic over-accumulation. Zinc uptake in S. cerevisiae is controlled at the transcriptional level in response to intracellular zinc concentrations (Zhao & Eide, Citation1996a, Citation1996b), and the regulation of these genes is mediated by the Zap1 transcription factor, which responds to zinc concentration regardless of the ambient pH (Zhao & Eide, Citation1997). However, other transcription factors involved in regulating ZIP transporter expression in Aspergillus fumigatus include the pH responsive PacC/Rim101p. ZRFA and ZRFB (ZRT1 and ZRT2 orthologs respectively) are also required for acidic zinc-limiting conditions growth, whereas they are dispensable for neutral or alkaline pH zinc-limiting media growth, being repressed by PacC transcriptional regulator (Amich, Citation2010).
In Ustilago maydis we found that UmZRT1 and UmZRT2 were expressed in acidic and neutral zinc-limiting media. At pH 4.0, UmZRT1 and NRG1 expression levels did not present considerable variations between FB1 and BMA2 (ΔRIM101) in our experimental conditions. Therefore, UmZRT1 is unlikely to be regulated by Rim101p or by Nrg1p. In contrast, UmZRT2 showed an ∼800-fold increased expression in BMA2 (), so its expression is likely regulated by Rim101p, as repressor of UmZRT2 at acidic pH and low zinc concentrations in the WT FB1 strain, either directly or indirectly by activating another regulator expression in this specific condition. This observation suggests that there could be an active form of PacC/Rim101p able to regulate its target genes not only at neutral/alkaline environment, but also under acidic conditions. This represents more evidence for Rim101/PacCp activity at acidic pH and it reinforces previous data when septum deposition has been observed at acid pH in U. maydis RIM101 mutants (Aréchiga & Ruiz-Herrera, Citation2005). Additionally, probable PacCp activity under acidic conditions in Trichoderma viridens was also reported (Trushina et al., Citation2013). It was expected that the ZRT2 ortholog in U. maydis would be expressed at a lower level than UmZRT1 at low zinc concentrations in acidic environments because Zrt1p is the primary system for the acquisition of Zn2+ at low concentrations of the metal in S. cerevisiae. Also in that yeast, Zrt1p accumulation is about 100-fold elevated (Zhao & Eide, Citation1996b) while Zrt2p is repressed to low expression levels in severe zinc deficiency, probably because of its low zinc affinity (Bird et al., Citation2004). In A. fumigatus, at neutral and alkaline pH, PacCp represses the expression of those genes expressed in acidic conditions, among them ZRFA/ZRFB (Amich, Citation2010). In contrast we found that in Ustilago maydis FB1, UmZRT2 expression is upregulated at neutral pH and low zinc concentration more than 1000-fold (), providing evidence of divergence with other systems previously reported in the acquisition of zinc.
Zinc is an essential metal for cell physiology and while few studies on Zn homeostasis are available for Basidiomycetes, our data provides an avenue to further Zn homeostasis studies in nontraditional fungi models. Our results also identify novel potential transcriptional regulators such as Rim101p at non-canonical conditions such as acidic pH. Further studies are required to assess if our observations can be extrapolated to other members of the phylum.
Supplemental Material
Download PDF (323.5 KB)Acknowledgement
Thanks are given to Roberto Mercado Hernández PhD for his advice in statistical methods applied in this work.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Adiloglu A, Adiloglu S. 2006. The effect of boron (B) application on the growth and nutrient contents of maize in zinc (Zn) deficient soils. Res J Agric Bio Sci 2:1–4.
- Alloway BJ. 2004. Zinc in Soils and Crop Nutrition. International Zinc Association Communications. Brussels, Belgium: IZA publications.
- Alloway BJ. 2008. Zinc in Soils and Crop Nutrition. 2nd ed. Brussels, Belgium and Paris, France: International Zinc Association (IZA) and International Fertilizer Association (IFA), 139.
- Alloway BJ. 2009. Soil factors associated with zinc deficiency in crops and humans. Environ Geochem Health 31:537–548.
- Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O. 2017. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 33:3387–3395.
- Amberg DC, Burke D, Strathern JN. 2005. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, New York, NY: Cold Spring Harbor Laboratory Press, 11–17.
- Amich J. 2010. Homeostasis del zinc a pH alcalino y su relevancia en la virulencia de Aspergillus fumigatus. [Zinc homeostasis at alkaline pH and its relevance in Aspergillus fumigatus virulence], [dissertation]. España: Universidad de Salamanca.
- Amich J, Leal F, Calera JA. 2009. Repression of the acid ZrfA/ZrfB zinc-uptake system of Aspergillus fumigatus mediated by PacC under neutral, zinc-limiting conditions. Int Microbiol 12:39–47.
- Amich J, Vicente-franqueira R, Mellado E, Ruiz-Carmuega A, Leal F, Calera J. 2014. The ZrfC alkaline zinc transporter is required for Aspergillus fumigatus virulence and its growth in the presence of the Zn/Mn-chelating protein calprotectin. Cell Microbiol 16:548–564.
- Aréchiga ET, Ruiz-Herrera J. 2005. The RIM101/PacC homologue from the Basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukaryot Cell 4:999–1008.
- Balaji RV, Colvin RA. 2005. A proton-dependent zinc uptake in PC12 cells. Neurochem Res 30:171–176.
- Banuett F, Herskowitz I. 1989. Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc Natl Acad Sci USA 86:5878–5882.
- Baslam M, Qaddoury A, Goicoechea N. 2014. Role of native and exotic mycorrhizal symbiosis to develop morphological, physiological and biochemical responses coping with water drought of date palm, Phoenix dactylifera. Trees 28:161–172.
- Bers DM, Patton CW, Nuccitelli R. 2010. A practical guide to the preparation of Ca2+ buffers. Methods in cell biology. Vol. 99. Cambridge, MA: Academic Press; 1–26.
- Biesalski HK. 2013. Hidden Hunger. Berlin, Heidelberg: Springer, 25–50.
- Bird A, Blankman E, Stillman D, Eide D, Winge R. 2004. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. Embo J 23:1123–1132.
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132.
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res 41:W349–W357.
- Cakmak I, Pfeiffer W, Mcclafferty B. 2010. Biofortification of durum wheat with zinc and iron. Cereal Chem 87:10–20.
- Catlett NL, Lee BN, Yoder OC, Turgeon BG. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Rep 50:9–11.
- Cavagnaro TR. 2008. The role of arbuscular mycorrhizas in improving plant zinc nutrition under low soil zinc concentrations: a review. Plant Soil 304:315–325.
- de Mattos-Shipley KM, Ford KL, Alberti F, Banks AM, Bailey AM, Foster GD. 2016. The good, the bad and the tasty: The many roles of mushrooms. Stud Mycol 85:125–157.
- Eide D. 2005. The Zip family of zinc transporters. Zinc Finger Proteins. Boston, MA: Springer, 261–264.
- Eide D. 2006. Zinc transporters and the cellular trafficking of zinc. Biochim Biophys Acta 1763:711–722.
- Eide D, Guarente L. 1992. Increased dosage of a transcriptional activator gene enhances iron-limited growth of Saccharomyces cerevisiae. J Gen Microbiol 138:347–354.
- Fairhead C, Llorente B, Denis F, Soler M, Dujon B. 1996. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using 'split-marker' recombination. Yeast 12:1439–1457.
- Fairhead C, Thierry A, Denis F, Eck M, Dujon B. 1998. 'Mass-murder' of ORFs from three regions of chromosome XI from Saccharomyces cerevisiae. Gene 223:33–46.
- Figueiredo DD, Barros PM, Cordeiro AM, Serra TS, Lourenço T, Chander S, et al. 2012. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J Exp Bot 63:3643–3656.
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230.
- Gaither L, Eide D. 2001. Eukaryotic zinc transporters and their regulation. BioMetals 14:251–270.
- Green MR, Sambrook J. 2012. Molecular Cloning: A Laboratory Manual, 4th edition. Cold Spring Harbor, New York, NY: Cold Spring Harbor Laboratory Press; 11–26, 362–372.
- Guerinot ML. 2000. The ZIP family of metal transporters. Biochim Biophys Acta 1465:190–198.
- Hamid A, Ahmad N. 2001. Paper at regional workshop on integrated plant nutrition system. (IPNS): development and rural poverty alleviation, 18–21 Sept, Bangkok.
- Hofmann K, Stoffel W. 1993. TMbase-A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 374:166.
- Holliday R. 1961. Induced mitotic crossing-over in Ustilago maydis. Genet Res 2:231–248.
- Hotz C, Brown KH. 2004. Assessment of the risk of zinc deficiency in populations and options for its control. Food Nutr Bull 25:94–204.
- Jin W, Peng L, Zhang X, Sun H, Yuan Z. 2019. Effects of endophytic and ectomycorrhizal basidiomycetes on Quercus virginiana seedling growth and nutrient absorption. J Sustain Forest 38:1–14.
- Kabala C, Singh BR. 2001. Fractionation and mobility of copper, lead, and zinc in soil profiles in the vicinity of a copper smelter. J Environ Qual 30:485–492.
- Kämper J, Kahmann R, Bölker M, Ma L, Brefort T, Saville BJ, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 44:97–101.
- Kramer U, Clemens S. 2005. Function and homeostasis of zinc, copper, and nickel in plants. Topics Curr Genet 14:215–271.
- Lamb TM, Xu W, Diamond A, Mitchell AP. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem 276:1850–1856.
- Lamb T, Mitchell AP. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol 23:677–686.
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408.
- MacDiarmid C, Gaither L, Eide D. 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. Embo J 19:2845–2855.
- Mendoza-Cózatl DG, Xie Q, Akmakjian GZ, Jobe TO, Patel A, Stacey MG, et al. 2014. OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol Plant 7:1455–1469.
- Mewes HW, Amid C, Arnold R, Frishman D, Güldener U, Mannhaupt G, et al. 2004. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res 32:41D–44.
- NCBI Resource Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44: (Database issue): D7–D19.
- Nishida S, Mizuno T, Obata H. 2008. Involvement of histidine-rich domain of ZIP family transporter TjZNT1 in metal ion specificity. Plant Physiol Biochem 46:601–606.
- Omasits U, Ahrens CH, Müller S, Wollscheid B. 2014. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886.
- Raklami A, Bechtaoui N, Tahiri AI, Anli M, Meddich A, Oufdou K. 2019. Use of Rhizobacteria and Mycorrhizae Consortium in the open field as a strategy for improving crop nutrition, productivity and soil fertility. Front Microbiol 10:1106.
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. 1995. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370:264–268.
- Ruiz-Herrera J. 2008. Ustilago maydis: ascenso de un hongo mexicano de la gastronomía local al mundo científico [Ustilago maydis: rise of a Mexican fungus from local gastronomy to the scientific world]. Nova Scientia 1:118–135.
- Serrano R, Ruiz A, Bernal D, Chambers JR, Arino J. 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol Microbiol 46:1319–1333.
- Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189:8746–8749.
- SPSS Inc. 2008. SPSS Statistics for Windows, Version 17.0. Chicago: SPSS Inc.
- Stein AJ, Nestel P, Meenakshi JV, Qaim M, Sachdev HP, Bhutta ZA. 2007. Plant breeding to control zinc deficiency in India: how cost-effective is biofortification? Public Health Nutr 10:492–501.
- Tapiero H, Tew KD. 2003. Trace elements in human physiology and pathology: zinc and metallothioneins. Biomed Pharmacother 57:399–411.
- Teixeira MC, Monteiro PT, Palma M, Costa C, Godinho CP, País P, et al. 2018. YEASTRACT: an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res 46:D348–D353.
- The UniProt Consortium. 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res 46:2699
- Trushina N, Levin M, Mukherjee K, Horwitz B. 2013. PacC and pH–dependent transcriptome of the mycotrophic fungus Trichoderma virens. BMC Genomics 14:138.
- Tusnády GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850.
- Vyas VK, Berkey CD, Miyao T, Carlson M. 2005. Repressors Nrg1 and Nrg2 regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot Cell 4:1882–1891.
- Velu G, Singh RP, Huerta-Espino J, Peña-Bautista RJ, Ortiz-Monasterio I. 2011. Breeding for enhanced zinc and iron concentration in CIMMYT spring wheat germplasm. Czech J Genet Plant Breed 47:(Special Issue): S174–S177.
- Welch R, Graham R. 2004. Breeding for micronutrients in staple food crops from a human nutrition perspective. J Exp Bot 55:353–364.
- Wilson D, Citiulo F, Hube B. 2012. Zinc exploitation by pathogenic fungi. PLoS Pathog 8:e1003034
- Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981.
- Zhao H, Butler E, Rodgers J, Spizzo T, Duesterhoeft S, Eide D. 1998. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J Biol Chem 273:28713–28720.
- Zhao H, Eide D. 1996a. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA 93:2454–2458.
- Zhao H, Eide D. 1996b. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem 271:23203–23210.
- Zhao H, Eide DJ. 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol Cell Biol 17:5044–5052.