Abstract
In this study, five secondary metabolites (caffeic acid, rosmarinic acid, lithospermic acid B, 12-hydroxyjasmonic acid 12-O-β-glucoside and p-menth-3-ene-1,2-diol 1-O-β-glucopyranoside) isolated from the polar extracts of the plant Origanum vulgare L. ssp. hirtum, were tested in vitro for their ability to inhibit soybean lipoxygenase. Among the examined compounds, lithospermic acid B demonstrated the best inhibitory activity on soybean lipoxygenase with IC50 = 0.1 mM. Docking studies have been undertaken as an attempt for better understanding the interactions of these compounds within the active site of soybean lipoxygenase. The predicted binding energy values correlated well with the observed biological data.
Introduction
Lipoxygenases (LOs) constitute a family of dioxygenases. They catalyze the oxygenation of free and esterified polyunsatured fatty acids containing a (1Z, 4Z)-penta-1,4-diene system to produce the corresponding hydroperoxy derivatives. They are monomeric proteins that contain a “non heme” iron per molecule in the active site as high spin Fe (II) in the native state and high-spin Fe (III) in the activated state [Citation1]. Their specificity depends on how the fatty acid is bound at the enzyme active site. LOs are categorized with respect to their positional specificity of arachidonic acid oxygenation. In theory, the following six LOs families may be distinguished: 5-LOs, 8-LOs, 9-LOs, 11-LOs, 12-LOs and 15-LOs[Citation2]. The process of dioxygenation as catalyzed by the enzyme, involves three consecutive steps: i) stereoselective hydrogen abstraction from a double allylic methylene group ii) radical rearrangement and iii) stereospecific insertion of molecular oxygen, reduction of hydroperoxy radical intermediate to the corresponding anion and release of lipid hydroperoxide LOOH. The products hydroperoxide of fatty acid, considered to be flavor precursors, are subsequently converted by several other enzymes into flavor compounds including ketones, aldehydes and alcohols [Citation3].
Lipoxygenases can be found in a wide variety of plants, fungi, and animal tissues. Eighteen carbon chain fatty acids (e.g. linoleic) are the primary substrates of the plant lipoxygenases, while the mammalian isozymes mainly catalyze the metabolism of fatty acids of carbon length 20 (e.g. arachidonic). Soybean lipoxygenase isozymes L1, L2, L3 have significantly different properties. L1 shows a much higher degree of regioselectivity and stereoselectivity in comparison with L3. The studies of the stereochemistry of the fatty acid hydroperoxide products of soybean lipoxygenase catalysis provide the evidence that the soybean isozymes differ in this aspect of catalysis. L1 acting on linoleic or arachidonic acid produces predominantely 13-hydroperoxy-9-cis,11-trans products, with 13R:13S approximately 10:90. L3 delivers a mixture (50:50) of 9- and 13-hydroperoxides showing a preference for cis-trans vs. trans-trans dienoic system (approximately 60:40) and with higher turnout of R stereoisomers over S (approximately 60:40) [Citation4].
The crystallographic data indicates that LOs have two domains: a smaller N-terminal β-barrel domain and a larger, helical catalytic domain toward the C-terminus containing a single atom of non-heme iron. The metal is connected as a ligand to conserved histidines and to the carboxyl group of a conserved isoleucine at the C-terminus of the protein. Several crystallographic determined structures of lipoxygenases (two isoforms from soybeans and one from rabbit) have been reported Citation5-8. The amino acid sequences between plant and mammalian LO enzymes show considerable homology. The soybean lipoxygenases, L1 and L3, are 72% identical in their amino acid sequences, but share only 25% sequence homology to any mammalian 15-LO. The highest level of sequence identity between lipoxygenases from plants and mammals lies in the area of the catalytic domain containing the non-heme iron atom [Citation9]. The only significant difference in the iron amino acid ligands is that in rabbit enzyme one of the iron coordination positions is occupied by histidine, while the structures of the soybean enzymes contain asparagines in the same location, but at just beyond a bonding distance [Citation7].
There is considerable evidence that lipoxygenase products are important mediators of the pathophysiology of asthma, heart diseases, cancer, inflammatory disorders [Citation10,Citation11]. In view of their pathophysiological properties, intervention with the biosynthesis or the action of leukotrienes (LTs) proposes a therapeutic benefit in a variety of allergic and inflammatory diseases. In regard to its characteristics and mechanism of action, different strategies have been developed to inhibit the LO pathway. Direct approaches involve: a) antioxidants and free radical scavengers, since lipoxygenation occurs via a carbon centered radical, b) iron ligand inhibitors, c) non redox competitive inhibitors, d) combination of being both chelators and reductants [Citation12].
From our previous study [Citation13] it was demonstrated that the polar extracts of Origanum vulgare L. ssp. hirtum on soybean lipoxygenase possessed inhibitory activity in vitro. Thus, we found it of interest to examine inhibition by the recently isolated polar secondary metabolites[Citation14]: caffeic acid (1), rosmarinic acid (2), lithospermic acid B (3), 12-hydroxyjasmonic acid 12-O-β-glucoside (4), and p-menth-3-ene-1,2-diol 1-O-β-glucopyranoside, of soybean lipoxygenase (5) ().
Figure 1 Chemical structures of the investigated compounds: caffeic acid (1), rosmarinic acid (2), lithospermic acid B (3), 12-hydroxyjasmonic acid 12-O-β-glucoside (4), and p-menth-3-ene-1,2-diol 1-O-β-glucopyranoside (5).
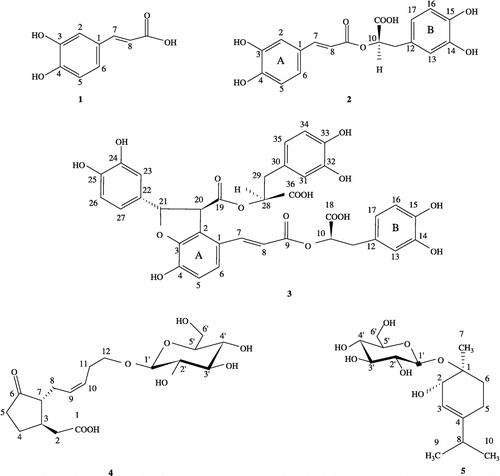
Compounds 1–3 have been examined for their inhibitory activity against 5-LOCitation15-18. However, all the tested compounds have been studied for their inhibitory activity on soybean LO for the first time in this study. Furthermore, we performed docking calculations via the GLUE program (implemented in the Grid-22 package) with the aim of explaining the differences in activity of our compounds. Understanding how these compounds interacts with soybean LO may explain how they inhibit lipoxygenases and LTs formation.
Materials and methods
Isolation and identification of the examined compounds
The compounds were extracted from the aerial parts of Origanum vulgare L. ssp. hirtum. The plant was collected from Pogoni-Ioannina (Epirus, North-Western Greece) in July 2002 and authenticated by Dr. Th. Constantinidis (Institute of Systematic Botany, Agricultural University of Athens). A voucher specimen is deposited in the Herbarium of the Institute of Systematic Botany, Agricultural University of Athens (ACA), Lazari 1.
The isolation and the structure elucidation of the isolated compounds have been described by the authors Koukoulitsa et al [Citation13].
Determination of soybean lipoxygenase inhibition in vitro [Citation19]
Soybean lipoxygenase and linoleic acid sodium salt were obtained from Sigma Chemical, Co. (St. Louis, MO, USA). Quercetin was purchased from the Aldrich Chemical Co. (Milwaukee, WI, USA). All reagents of the in vitro experiment were obtained from commercial sources and were of analytical grade. Each experiment of the in vitro assay was performed at least in triplicate and the standard deviation of absorbance was less than 10% of the mean. The examined compounds dissolved in DMSO, were incubated at room temperature with 0.1 mL sodium linoleate (10− 4 M), and 0.2 mL of enzyme solution (0.11 × 104, w/v in saline) in pH 9 (Tris as a buffer) with final volume 1 mL. The conversion of sodium linoleate to 13-hydroperoxylinoleic acid at 234 nm after 3 min was recorded; IC50 values ( ± s.d.) were determined for each compound and compared with an appropriate standard inhibitor ().
Table I. Inhibition of soybean lipoxygenase in vitro IC50 (mM), calculated binding energy for the highest ranked docking solution and calculated Log P value of the docked conformers.
Computational methods
Docking calculations
The studied molecules were built in 3D coordinates and their most stable (lower energy) conformations were calculated, by geometrical optimization of their structure as implemented in the Spartan '04 Molecular Modeling program suite [Citation20] utilizing MMFF94 molecular mechanics force field and PM3 semi-empirical method. The X-ray structure of human soybean lipoxygenase (PDB entry code: 1IK3) was used in our docking calculations after deletion of 13-HPOD from the PDB file, obtained from the Brookhaven Protein Data Bank [Citation21].
Docking calculations were performed with the aid of GLUE program implemented in the GRID package (www.moldiscovery.com) [Citation22]. GLUE [Citation23] is a docking procedure aimed at detecting energetically favorable binding modes of a ligand with respect to the protein active site using the GRID force field [Citation24].
Results and discussion
Biological results
The studied compounds, 1–5, were evaluated for their inhibitory activity against soybean lipoxygenase in vitro. All the tested compounds were found to exhibit sufficient inhibitory activity (). Lithospermic acid B (3) was the most potent compound (IC50 = 0.10 mM), whereas rosmarinic acid (2) presented the lowest activity under the reported experimental conditions. The inhibitory activity of the remained compounds followed the order: p-menth-3-ene-1,2-diol 1-O-β-glucopyranoside>12-hydroxyjasmonic acid 12-O-β-glucoside>caffeic acid.
It is known that lipophilicity is an important property for the inhibition of LO [Citation27]. From our results it is confirmed that compound 3 with the highest Log P value (calculated by VolSurf [Citation25,Citation26]) () presents the higher inhibitory activity, but this was not followed by the remaining compounds.
Docking studies of receptor-ligand interactions
Due to the lack of structural data for human LOs, researchers are still modeling human LOs using soybean enzymes because of their availability and highly characterized structures. Use of plant lipoxygenases to model mammalian LOs will prove highly beneficial and supportive in structural characterization, mechanism elucidation, and possibly the discovery of novel inhibitors of LOs [Citation5].
The possible mechanism of action, as well as the differences in activity of our compounds toward soybean lipoxygenase could be explained by docking calculations. Since the structures of the native L3 and the complex present few differences, either in the protein or in the solvent, for the docking study was selected isozyme L3 complexed with 13-HPOD [9Z,11E-13(S)-hydroxyperoxy-9,11-octadecadienoic acid] (a product of linoleic acid degradation) (). For the docking study 13-HPOD, a product of linoleic acid degradation, was selected[Citation9]. The X-ray crystallography study revealed that the substrate anchors its carboxyl group to the hydrophilic region lined by residues Ser510 and Arg726, Gln716, Asp766 and Gly720. The 9-cis, 11-trans double bond system has His518-Trp519 on one side and the hydrophobic residues Leu565, Ile572 and Leu773 on the opposite side in the immediate vicinity. The aliphatic end of the molecule, C14–C18, is squeezed between Ile857 and Leu277 protruding into a hydrophobic channel, lined by the side chains of Leu273, Thr274, Leu277, Ile557, Ile857 and Ile772 [Citation7].
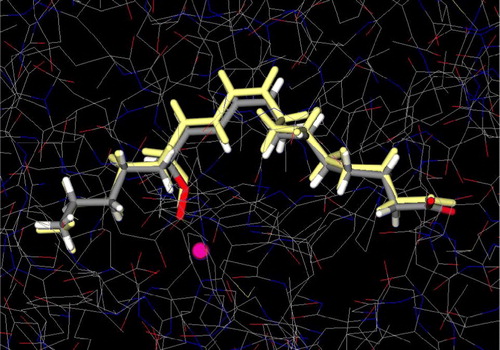
The docking simulations in the active site of L3 were performed by the docking program GLUE, which has been shown to successfully reproduce experimentally observed binding modes, in terms of r.m.s.d [Citation23,Citation28]. The energy-minimized structure of the ligand was docked into L3 and GLUE provided excellent result as it was observed to give a low value of r.m.s.d (best docked solution 0.36 Å) between the experimental structure and the calculated docked structure (). The ability to accurately predict the binding conformation of the ligand gave confidence that GLUE would exhibit a similar accuracy with the investigated molecules utilized in the study.
Figure 3 Conformational comparison between X-ray structure of 13-HPOD (colored by atom type) (PDB IK3) and predicted (colored yellow) by GLUE. Iron atom is illustrated as a sphere (colored magenta). Colour plate can be viewed in the online version.
In are illustrated the best possible binding modes of the molecules at the active site of L3. Their corresponding binding energies are shown in . As can be seen, the molecules are positioned in the same place occupied by the substrate. The enzyme/inhibitor complexes are stabilized almost entirely by van der Waals and aromatic interactions in the hydrophobic region and by hydrogen bonds in the hydrophilic region.
Figure 4 Docked orientations of a. caffeic acid (1) b. rosmarinic acid (2) c. lithospermic acid B (3) d. 12-hydroxyjasmonic acid 12-O-β-glucopyranoside (4) and e. p-menth-3-ene-1,2-diol 1-O-β-glucopyranoside (5) (illustrated as a ball-and-stick model colored by atom type) with additional depiction of selected amino acid residues of soybean lipoxygenase (PDB IK3) as well as 13-HPOD (drawn as sticks). Hydrogen bonds and polar interactions are shown as dotted lines. White colored circles highlight double bond position of the compounds. Iron atom is illustrated as a sphere (colored magenta). Coloured plate can be viewed in the online version.
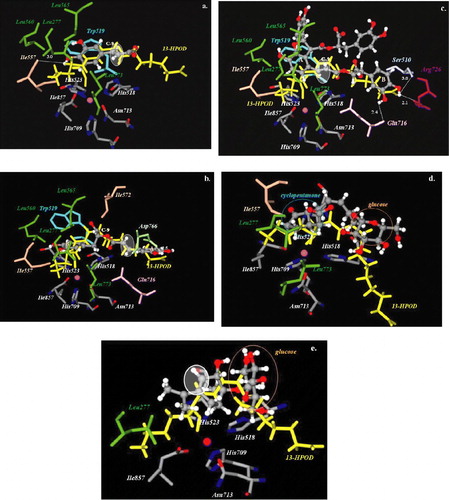
In particular, lithospermic acid B (3) penetrated into the central cavity with its trans C7–C8 double bond positioned near the cis C9–C10 double bond of the substrate. The molecule was stabilized by hydrophobic contacts of phenyl ring A through residues Ile557, Leu277, Ile857, Leu773 and by aromatic interactions with Trp519. Further stabilization of the molecule was attributed to the hydrogen bonds formed between hydroxyl groups of phenyl ring B and the residues of the hydrophilic region Gln716, Ser510, and Arg726 (2.4 Å, 3.0 Å, and 2.1 Å respectively) ().
Caffeic acid (1) was also positioned at the active site with its double bond near the cis C9–C10 double bond of the substrate. Hydrophobic interactions involving the residues Leu560, Leu277, Leu565, Leu773, Ile557, Ile857 and aromatic interactions with Trp519 were observed. Moreover, a hydrogen bond was observed between phenyl hydrogen and Ile557 (3.0 Å) ().
Rosmarinic acid (2) entered into the hydrophilic region of the enzyme more deeply than caffeic acid and its double bond is not in proximity with the double bond of the substrate. Phenyl ring A forms a hydrogen bond with Asp766 (2.5 Å) through its hydroxyl groups at C-3. Hydrophobic contacts were established between phenyl ring B and the amino acids of the hydrophobic region ().
Compounds 4 and 5 are characterized by both hydrophobic and glucose moieties. As depicted in and , the hydrophobic groups of these compounds are oriented towards the hydrophobic area of the active site, whereas the glucose moieties are oriented towards the hydrophilic area. The position of the double bond of these compounds was observed to be adjacent to the substrate's double bond.
The docking study suggests that the biological response of the examined compounds could be explained by the proper geometry of the predicted conformations at the active site. As is depicted in , a decrease in binding energy of the examined compounds reflects an increase in their inhibitory activity. Regarding compounds 1 and 3–5, their double bonds as well as the double bond of the substrate are in close proximity (, 4c–4e). As a result, the molecules are able to mimic the role of the double bond of the substrate and in this way exhibit their inhibitory activity. For compounds 1 and 4–5 similar binding poses may explain their binding energies and the observed inhibitory activities. Compound 3, which was found to be the stronger inhibitor (IC50 = 0.10 mM) also exhibits the lowest binding energy ( − 19.52 Kcal/mol). On the contrary, as it is illustrated in , rosmarinic acid's double bond is not adjacent to the double bond of the substrate. The highest observed binding energy for rosmarinic acid ( − 4.93 Kcal/mol) compared to those of the remaining compounds ( − 9.70 Kcal/mol to − 19.52 Kcal/mol) () is in accordance with its weaker inhibitory activity. From the above results, it was deduced that the predicted binding energy values correlated well with the observed biological data.
Conclusion
The inhibitory activity against soybean lipoxygenase of compounds 1–5 was investigated in vitro and docking studies at the active site of the enzyme were performed. Lithospermic acid B was found to be the most active compound. Docking results significantly support the in vitro biological data.
| Abbreviations | ||
| LOs: | = | lipoxygenases |
| LTs: | = | leukotrienes |
Acknowledgements
We are grateful to Prof. Gabriele Cruciani (Laboratory for Chemometrics, School of Chemistry, University of Perugia, Italy) for providing us GRID package and VolSurf program. We also thank Dr Simone Sciabola for useful advices and Dr Paolo Benedetti and Dr Giuliano Berellini for technical assistance (same laboratory).
References
- Boyington JC, Amzel L. J Biol Chem 1990; 265: 12771–12773
- Kuhn H. Prostag. Lipid Mediat 2000; 62: 255–270
- Nanda S, Yadav JS. J Mol Cat B: Enzymatic 2003; 26: 3–28
- Skrzypczak-Jankun E, Amzel LM, Kroa BA, Funk MO. Proteins: Struct Funct Genet 1997; 29: 15–31
- Minor W, Steczko J, Stec B, Otwinowski Z, Bollin JT, Walter R, Axelrod B. Biochemistry 1996; 35: 10687–10701
- Tomchick DR, Phan P, Cymborowski M, Minor W, Holman TR. Biochemistry 2001; 40: 7509–7517
- Skrzypczak-Jankun E, Bross RA, Caroll RT, Dunham WR, Funk MO. J Am Chem Soc 2001; 123: 10814–10820
- Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. Nat Struct Biol 1997; 4: 1003–1009
- Skrzypczak-Jankun E, McCabe NP, Selman SH, Jankun J. Int J Mol Med 2000; 6: 521–526
- Wenzel SE. Pharmacother 1997; 17:3S–12S.
- Steele VE, Holmes CA, Hawk ET, Kopelovich L, Lubet RA, Crowell JA, Sigman CC, Kelloff GJ. Cancer Epidemiol Biomarkers Prev 1999; 8: 467–483
- Kulkarni AP. Cell Mol Life Sci 2001; 58: 1805–1825
- Koukoulitsa C, Zika C, Hadjipavlou-Litina D, Demopoulos VJ, Skaltsa H. Phytother Res 2006; 20: 605–606
- Koukoulitsa C, Karioti A, Bergonzi CM, Pescitelli G, Di Bari L, Skaltsa HJ. Agr Food Chem 2006; 54: 5388–5392
- Koshihara Y, Neichi T, Murota S, Lao A, Fujimoto Y, Tatsuno T. Biochim Biophys Acta 1984; 792: 92–97
- Sugiura M, Naito Y, Yamaura Y, Fukaya C, Yokoyama K. Chem Pharm Bull 1989; 37: 1039–1043
- Aoshima Jiro, Arai Junichiro, Isumi Kazuhiro, Kusunoki Shinichiro. Jpn Kokai Tokkyo Koho 1989; p.3, CA 111:225310.
- Tomita Yutaka, Ikeshiro Yasumasa, Ushijima Hideto, Wakabayashi Toshio. Jpn. Kokai Tokkyo Koho 1990; p.4, CA 114:69040.
- Kontogiorgis C, Hadjipavlou-Litina D. J Enz Inhib Med Chem 2003; 18: 63–69
- Spartan ′04 Windows., Wavefunction, Inc., Irving, CA.
- RCSB Protein Data Bank, operated by the Research Collaboratory for Structural Bioinformatics.
- The version 22 of the GRID package is available from Molecular Discovery Ltd., 4., Chandos Street, W1A 3BQ, London. United Kingdom.
- Carosati E, Sciabola S, Cruciani G. J Med Chem 2004; 47: 5114–5125
- Goodford PJ. J Med Chem 1985; 28: 849–857
- VolSurf is available from Molecular Discovery Ltd., 4, Chandos Street, W1A 3BQ, London. United Kingdom.
- Cruciani G, Pastor M, Guba W. Eur J Pharm Sci 2000; 11(Suppl. 2)S29–S39
- Pontiki E, Hadjipavlou-Litina D. Mini Rev Med Chem 2003; 3: 487–499
- Sciabola S, Carosati E, Baroni M, Mannhold R. J Med Chem 2005; 48: 3756–3767
![Figure 2 Chemical structure of 13-HPOD [9Z,11E-13(S)-hydroxyperoxy-9,11-octadecadienoic acid].](/cms/asset/b2ebb704-0330-475f-b83a-db63aeb4a2a3/ienz_a_199002_f0002_b.gif)