Abstract
Two new pentacyclic triterpenes eleganene-A (1) and eleganene-B (2), along with four known pentacyclic triterpenes betulin (3), ursolic acid (4), erythrodiol (5) and corosolic acid (6) were isolated from the aerial parts of Myricaria elegans. These compounds exhibited significant antibacterial activity. The structure of compounds 1 and 2 were deduced on the basis of their spectral analysis.
Introduction
Myricaria elegans Royle. (tamariscaceae) is a bush with a straight slender stem [Citation1]. The plant is found in western Himalaya, western Tibet and from Garwhal to Ladak at altitude of 6000–15,000 feet [Citation2]. The leaves of the plant are applied to the bruises and twigs are browsed by sheep and goats I Ladak [Citation3]. No phytochemical work was reported on this medicinal plant till today.
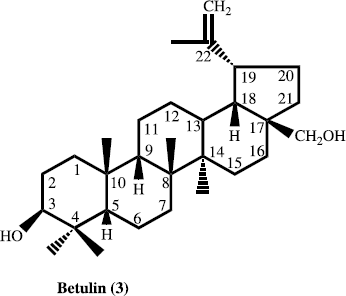
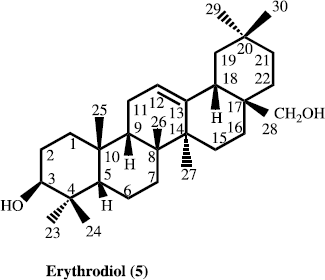
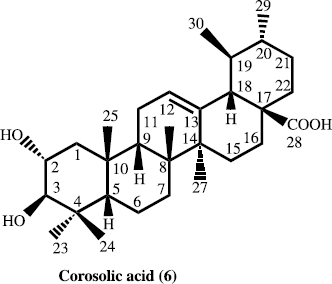
In the present paper, we describe the isolation and structure elucidation of two new antibacterial pentacyclic triterpene eleganene-A (1) and eleganene-B (2), along with four known new source pentacyclic triterpen betulin (3), ursolic acid (4), erythrodiol (5) and corosolic acid (6) for the first time from M. elegans.


Experimental
General
Optical rotations were measured on a JASCO DIP 360 polarimeter. IR spectra were recorded on a JASCO 302-A spectrophotometer. EI-MS and HREI-MS were recorded on JMS HX 110 with data system and on JMS-DA 500 mass spectrometers. The 1H- and 13C-NMR spectrums were recorded on Bruker NMR spectrometers operating at 400 MHz, (100 and 125 MHz for 13C). The chemical shifts values are reported in ppm (δ) units and the coupling constants (J) are given in Hz.
Chromatographic conditions
For TLC, precoated aluminium sheets (silica gel 60F-254, E. Merck) were used. Visualization of the TLC plates was achieved under UV at 254 and 366 nm and by spraying with cerric sulphate reagent. Solvent system; “n-hexane-ethylacetate 8:2”, was used.
Plant material
The aerial parts (5 kg wt) of Myricaria elegans Royle., were collected from District Swat of Pakistan at an elevation of 6000 feet in July 2002 and identified by Mr. Mehboob ur Rahman (Assistant Professor) Department of Botany, Jahanzeb Post Graduate College, Saidu Sharif, Swat, NWFP, Pakistan. The voucher specimen (EM-004) is deposited in the herbarium of the botany department.
Extraction and isolation
The air-dried aerial parts (5 kg) of M. elegans were grounded, dried and soaked in a mixture of 30 L of 80% MeOH/H2O (4:1) at room temperature for 72 h. The organic extract was filtered and solvent was evaporated under reduced pressures. The concentrated methanolic extract (560.3 g) was suspended in distilled water (1.5 L) and defatted with pet. ether (2.5 L). The aqueous layer was further sequential extracted with chloroform, ethyl acetate and butanol. Each extract was concentrated in vacuo to obtain pet. ether (55.2 g), chloroform (17.3 g), ethyl acetate (8.11 g) and butanol-soluble (52.5 g) fractions. The chloroform extract (17.3 g) was chromatographed on a silica gel column and eluted with n-hexane with gradient of polarity of chloroform upto 100%, and methanol upto 20%. Thus five sub-fractions were obtained. Sub-fraction ME-2, on repeated flash chromatography using 90% chloroform-methanol (90:10), as eluent afforded compound 1 (5.7 mg), and compound 2 (6.7 mg). Similarly from the sub-fraction ME-4, we got compound 3, 4 and 5, while, sub-fraction ME-5 afforded compound 6.
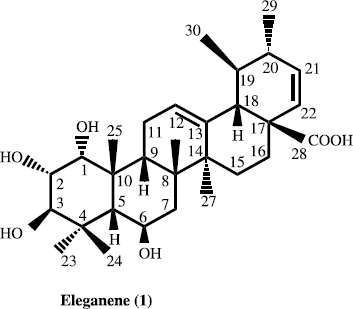
Eleganene-A (1)
White amorphous powder (5.7 mg); mp 180–182° C; [α]D 29+84° (c 0.8, CHCl3); UV (MeOH) λmax (log ε) 202 nm; IR (CDCl3) γmax 3665, 3363 (hydroxyl groups) and 1720 (carbonyl) cm− 1; 1H-NMR (400 MHz, CDCl3): δ 4.17 (H-1), δ 3.76 (H-2), δ 3.93 (H-3), δ 1.79 (H-5), δ 4.17 (H-6), δ 1.59 (H-7), δ 1.78 (H-9), δ 2.39 (H-11), δ 5.56 (H-12), δ 1.94 (H-15), δ 1.78 (H-16), δ 3.10 (H-18), δ 1.59 (H-19), δ 1.93 (H-20), δ 6.00 (H-21), δ 6.21 (H-22), δ 1.18 (CH3- 27), δ 1.08 (CH3- 23), δ 1.03 (CH3-24), δ 1.11 (CH3-26), δ 0.99 (CH3-29), δ 0.95 (CH3-30), and δ 0.87 (CH3-25). 13C-NMR (CDCl3, 100 MHz): see : EIMS m/z (rel. int. %) 502 (2) [M]+, 484 (2.1), 468 (2.1), 288 (5.4), 248 (100), 233 (10.5), 190 (10.3), 173 (12.2), 133 (24.3), 119 (15.8), 105 (10.5). FAB + ve m/z 503; HR-EIMS m/z 502.30635 (calc. 502.30829 for C30H46O6).
Table I. 13C–NMR data of compound 1 and compound 2 (CDCl3).
Eleganene-B (2)
White amorphous powder (6.7 mg); mp 230–234° C; [α]D 29+74° (c 0.9, CHCl3); UV (MeOH) λmax (log ε) 202 nm; IR (CDCl3) γmax 3665, 3363 (hydroxyl groups) and 1705 (carbonyl) cm− 1; 1H-NMR (400 MHz, CDCl3): δ 5.61 (H-1), δ 5.50 (H-2), δ 3.89 (H-3), δ 1.03 (H-5), δ 1.72 (H-6), δ 1.31 (H-7), δ 1.95 (H-9), δ 1.99 (H-11), δ 5.26 (H-12), δ 1.50 (H-15), δ 1.93 (H-16), δ 2.33 (H-18), δ 1.49 (H-19), δ 1.63 (H-20), δ 1.44 (H-21), δ 1.91 (H-22), δ 1.27 (CH3- 27), 1.24 (CH3- 23), 1.04 (CH3-24), 1.01 (CH3-26), 0.99 (CH3-29), 0.95 (CH3-30), and δ 0.88 (CH3-25). 13C-NMR (CDCl3, 100 MHz): see : EIMS m/z (rel. int. %) 470 (2) [M]+, 456 (2.7), 447 (100), 219 (13.2), 207 (36.2), 203 (72.9), 189 (18.7), 175 (11), 133 (45), 119 (13.8), 105 (9.5), 67 (9.1), 55 (7.7). FAB + ve m/z 471; HR-EIMS m/z 470.20546 (calc. 470.20724 for C30H46O4).
Antibacterial activity
All of the isolated compounds were screened against strains of Escherichia coli, Bacillus subtilis, Shigella flexneri, Staphylococcus aureus, Pseudomonas aeruginosa, and Salmonella typhi. For antibacterial screening, 3 mg of sample was taken and dissolved in 3 mL of DMSO. Molten nutrient agar (45 mL) was poured on sterile petri plates, where it was allowed to solidify. Bacterial lawn were made on these nutrient agar plates by dispensing 7 mL of sterile soft agar containing 100 μL of test-organism culture. Wells were dug out with the help of a 6-mm sterile metallic borer at appropriate distance. Then, 10 mL of sample was poured into each well, and the plates were incubated at 37°C for 24 h. The results, in terms of inhibition zone, were noted. The drug Imipenem, a broad-spectrum β-lactam antibiotic, was used as a positive control. As a negative control, DMSO was used. The results of these experiments are summarized in .
Table II. Antibacterial activities of compounds 1–6. Relative to the standard drug imipenem.
Results and discussion
Chemistry
Eleganene-A (1) was obtained as a white amorphous powdered, from the chloroform extract of the M. elegans., in the IR spectrum of compound 1 displayed the absorption bands at 1692 and 3408 cm− 1, indicating the presence of carbonyl and hydroxyl groups, respectively. The EIMS spectrum of compound 1 showed the ion at m/z 484 [M+-18] indicating the loss of a water molecule. The molecular ion was deduced from [M + H]+ (m/z 502.30635) in the HR-EIMS and was assigned the molecular formula C30H46O6 (calcd. 502.30829). The mass was further confirmed by FAB + ve and FAB − ve m/z 503 and 501. This showed eight degree of unsaturation which accounting for five rings, two olefinic bonds and carbonyl carbon functionality. The base peak at m/z 246.3053 (D/E ring, [C16H22O2]) may arise by the retro Diels-Alder cleavage ring C, which indicated the presence of Δ12- Ursane skeleton with carboxylic group and one olefinic bond on ring D [Citation4].
The 1H-NMR spectrum of eleganene-A (1) exhibited signals for seven methyl protons δ 1.18 (CH3-27), 1.08 (CH3-23), 1.03 (CH3-24), 1.11 (CH3-26), 0.99 (CH3-29), 0.95 (CH3-30), and δ 0.87 (CH3-25). A characteristic downfield triplet resonating at δ 5.53 (1H, t, J = 2.4 Hz, H-12) was assigned to the olefinic C-12 proton. Similarly, in the downfield region a doublet of one proton integration at δ 6.21 (1H, t, J = 5.6 Hz, H-22) was assigned to the C-22 olefinic proton and a triplet of one proton at δ 6.00 (1H, t, J = 7.5 Hz, H-21) was assigned to the C-21 olefinic proton. While a doublet of two protons at δ 4.39 (2H, d, J = 10.5 Hz, H-1, 6) were assigned to C-1 and C-6 methine protons. In the spectrum a downfield broad triplet resonating at δ 4.22 (1H, t, J = 13.9 Hz, H-2) was assigned to the C-2 proton. While a doublet of one proton integration at δ 4.04 (1H, d, J = 10.4, H-3) was assigned to the C-3 methin protons geminal to hydroxyl group.
The broad band decoupled 13C-NMR spectrum (DEPT) (), showed the signals for thirty carbons including seven methyls, four methylene, twelve methine, and seven quaternary carbons. The 1H- 13C correlation was determined by the HMQC spectrum, while the long range 1H- 13C connectivities were obtained through HMBC technique.
Thus on the basis of above spectral data, the structure of compound 1 was deduced as eleganene-A (1).
Eleganene-B (2) was obtained as a white amorphous powdered, from the chloroform extract of the M. elegans. In the IR spectrum of compound 2 displayed the absorption bands at 1692 and 3408 cm− 1, indicating the presence of carbonyl and hydroxyl groups, respectively. The EIMS spectrum of compound 2 showed the ion at m/z 452 [M+-18] indicating the loss of a water molecule. The molecular ion was deduced from [M + H]+ (m/z 470.20546) in the HR-EIMS and was assigned the molecular formula C30H46O4 (calcd. 470.20724). The mass was further confirmed by FAB + ve m/z 471. This showed eight degree of unsaturation which accounting for five rings, two olefinic bonds and carbonyl carbon functionality. The base peak at m/z 248.4038 (D/E ring, [C16H24O2]) may arise by the retro Diels-Alder cleavage ring C, which indicated the presence of Δ12- Ursane skeleton with carboxylic group.
The 1H-NMR spectrum of eleganene-B (2) exhibited signals for seven methyl protons at δ 1.27 (CH3-27), 1.24 (CH3-23), 1.04 (CH3-24), 1.01 (CH3-26), 0.99 (CH3-29), 0.95 (CH3-30), and δ 0.88 (CH3-25). A characteristic downfield triplet resonating at δ 5.26 (1H, t, J = 3.4 Hz, H-12) was assigned to the olefinic C-12 proton. Similarly, in the downfield region a doublet of one proton integration at δ 5.61 (1H, d, J = 6.8 Hz, H-1) was assigned to the C-1 olefinic proton and a triplet of one proton at δ 5.50 (1H, t, J = 7.5 Hz, H-2) was assigned to the C-2 olefinic proton. While a doublet of one proton integration at δ 3.89 (1H, d, J = 10.4 Hz, H-3) was assigned to the C-3 methin protons geminal to hydroxyl group.
The broad band decoupled 13C-NMR spectrum (DEPT) (), showed the signals for thirty carbons including six methyls, eight methylene, nine methine, and seven quaternary carbons. The 1H- 13C correlation was determined by the HMQC spectrum, while the long range 1H- 13C connectivities were obtained through HMBC technique.
Thus on the basis of above spectral data, the structure of compound 2 was deduced as eleganene-B (2).
Antibacterial activity
Compounds 1 and 2 showed significant antibacterial activity against S. aureus. Compounds 3 and 4 showed significant activities against S. typhi and P. aeruginosa, while compounds 5 and 6 showed significant activity against S. typhi and P. aeruginosa ().
Acknowledgement
We are thankful to Prof. Mehboob-ur-Rahman for helping in the collection and identification of the plant.
References
- JD Hooker. Flora of British India. Vol. VI. Bishen Singh Mahendra Pal Singh, 23 A, New Connaught Dehra Dhun; (1982). 249–250.
- Kiritkar, and BD Basu. Indian medicinal plants. Allahabad: Sundhindra Nath Basu, Bahadur Ganj; (1918). p 141.
- G Watt. The dictionary of economic products of India. Vol. V. Part I. New Delhi: Cosmo publications; (1972). p310–311.
- H Budzikiewicz, JM Wilson, and C Djerassi. (1963). J Amer Chem Soc 85:3688.
