Abstract
Berberine was investigated as an inhibitor of human dipeptidyl peptidase IV (DPP IV) in an attempt to explain its anti-hyperglycemic activities. The investigation included simulated docking experiments to fit berberine within the binding pocket of DPP IV. Berberine was found to readily fit within the binding pocket of DPP IV in a low energy orientation characterized with optimal electrostatic attractive interactions bridging the isoquinolinium positively charged nitrogen atom (berberine) and the negatively charged acidic residue of glutamic acid-205 (GLU205) of DPP IV. Experimentally, berberine was found to inhibit human recombinant DPP IV in vitro with IC50 = 13.3 μM. Our findings suggest that DPP IV inhibition is, at least, one of the mechanisms that explain the anti-hyperglycemic activity of berberine. The fact that berberine was recently reported to potently inhibit the pro-diabetic target human protein tyrosine phosphatase 1B (h-PTP 1B) discloses a novel dual natural h-PTP 1B/DPP IV inhibitor.
| Abbreviations: | ||
| DPP IV, | = | Dipeptidyl peptidase IV |
| h-PTP 1B, | = | Human protein tyrosine phosphatase 1B |
| PBS, | = | Phosphate buffer saline |
| T2D, | = | Type 2 diabetes mellitus |
Introduction
Type 2 diabetes mellitus is a multifactorial and polygenic disease by which several organs are affected. Sedentary lifestyles, dietary changes and genetics are some of the key factors that have conspired to create the current worldwide epidemic of type 2 diabetes (T2D), an acquired syndrome of elevated blood glucose. An estimated 246 million people worldwide have T2D and this number is projected to grow to 366 million by 2030 [Citation1]. T2D accounts for more than 90% of diabetic cases [Citation2].
It is widely accepted that hyperglycemia in type 2 diabetes arises as a result of pancreas β-cell dysfunction, deficiency in insulin secretion, insulin resistance and/or increased hepatic glucose production. More recently, the role of other glucoregulatory hormones, including glucagon, amylin, and the gut peptides; glucagon-like peptide (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP); have also been included as important pathophysiologic defects in those with type 2 diabetes. GLP-1 and GIP are incretins released from the gut in response to food and play an essential role in maintaining glucose homeostasis [Citation3,Citation4]. Together, they are responsible for up to 70% of insulin secreted following a meal [Citation5-Citation8]. In patients with type 2 diabetes the incretin effect is known to be reduced largely because GLP-1 secretion is impaired, and this contributes to the inappropriately low insulin levels following oral ingestion of nutrients in these patients [Citation9,Citation10]. Continuous intravenous infusion of GLP-1 could normalize blood glucose concentration in these subjects [Citation11]. However, the effects of subcutaneous injections were short-lasting, and this was later shown to be due to the rapid degradation and inactivation of the peptide by the enzyme dipeptidyl peptidase IV (DPP IV) [Citation12]. DPP IV is a membrane-bound, serine protease ectoenzyme found in numerous sites, including the kidney, intestine, and capillary endothelium [Citation13]. It cleaves an NH2-terminal dipeptide from peptides where the penultimate amino acid residue is praline or alanine [Citation13]. DPP IV is responsible for the degradation of a number of biological peptides including GLP-1 and GIP [Citation14]. Consequently, inhibition of DPP IV has been shown to improve glucose tolerance in diabetic patients by enhancing the insulinotropic effects of GLP-1 [Citation15]. Moreover; DPP IV inhibitors promotes glucose uptake in cultured cells, induces ß-cell regeneration and inhibits apoptosis of ß-cells [Citation16, Citation17]. DPP-IV inhibition might offer advantages over existing therapies. For example, GLP-1 operates in a glucose-dependent manner, therefore there is a reduced risk of hypoglycemia [Citation18]. Moreover, it stimulates proliferation of insulin-producing β-cells providing additional benefit for diabetic patients. As a result, the search for inhibitors of DPP IV for the treatment of type 2 diabetes is an active area of research.[Citation19].
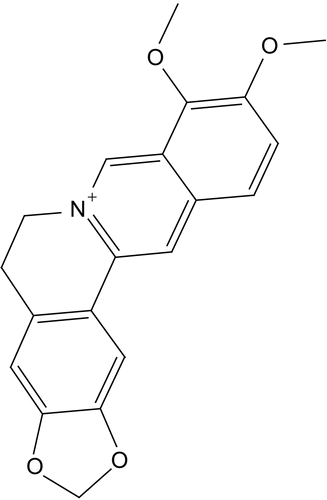
Berberine () is the main active component of an ancient Chinese herb Coptis chinensis French, which has been used to treat diabetes for thousands of years. Berberine is an isoquinoline alkaloid widely used in traditional eastern homeotherapy, particularly in treating gastrointestinal infections [Citation20-Citation22]. It has been extensively reviewed in the literature as interesting natural compound of wide potential medicinal applications [Citation23-Citation28].
Berberine has been reported to possess potent anti-diabetic activities [Citation29-Citation33]. The hypoglycemic effects of berberine were accidentally discovered when it was administered to a diabetic patient with diarrhea [Citation34]. Since then, berberine has often been used as an anti-hyperglycemic agent by many Chinese physicians.
Berberine has been shown to antagonize the hyperglycemic action of glucose and the gluconeogenic action of alanine in mice and hepatocyte cell lines [Citation32]. It seems to improve insulin resistance by raising insulin sensitivity [Citation31,Citation35]. In a study conducted on streptozocine-treated rats, berberine was found to decrease fasting blood glucose level and improve oral glucose tolerance, which was related to the property of stimulating insulin secretion [Citation29]. Interestingly, some recent reports suggested that berberine might inhibit the intestinal absorption of glucose [Citation36]. Nevertheless, most accumulated scientific evidence suggests that the hypoglycemic action of berberine is related to post-insulin receptor mechanisms [Citation29–32,Citation35]. Recently, our laboratory reported that berberine potently inhibits protein tyrosine phosphatase 1B (h-PTP 1B) (Ki value = 91.3 nM), which could partially explain its antidiabetic activity [Citation37].
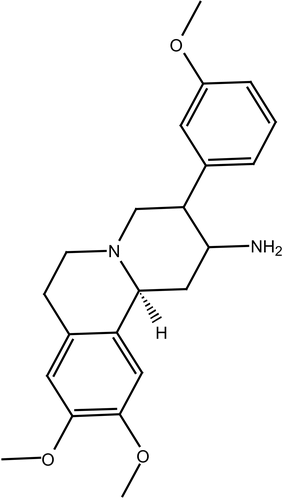
The anti-hyperglycemic action observed for berberine, combined with the observed similarity between berberine and the benzoquinolizines, a well known class of DPP IV inhibitors (e.g., compound 1, IC50 = 5.13 μM, ) [Citation38], prompted us to evaluate possible binding interaction(s) between berberine and DPP IV as a new suggested mechanism for the hypoglycemic action of this interesting natural compound.
We decided to evaluate the binding interactions by employing computer-aided molecular docking and scoring. Eventually, the findings of the docking simulation study were experimentally validated by an in vitro bioassay against human recombinant DPP IV.
Simulated molecular docking is basically a conformational sampling procedure in which various docked poses/conformations are explored to identify the right one. This process can be a very challenging problem given the degree of conformational flexibility at the ligand-macromolecular level [Citation39-Citation44]. Docking consists of two parts, namely, (i) prediction of the conformation, orientation and position (pose) of the bioactive compound into the binding pocket, and (ii) estimation of the tightness of target-ligand interactions (scoring) to guide conformational sampling [Citation45]. The final docked conformations are selected according to their scores. It was decided to conduct the docking study utilizing the program FRED [Citation46] which was recently reported to illustrate good overall performance, particularly in virtual high-throughput screening experiments (vHTS) [Citation47,Citation48].
Materials and methods
Materials
All of the chemicals used in these experiments were of molecular grades and obtained from commercial suppliers: H-Gly-Pro-para-Nitroaniline (BIOMOL, Germany), assay buffer components (Tris 50 mM) (BIOMOL, Germany), recombinant DPP IV (BIOMOL, Germany), Berberine (SIGMA, USA), DMSO (SIGM, USA), and standard DPP IV inhibitor (P32/98, BIOMOL, Germany).
Molecular modeling
Hardware and software
The following software packages were utilized in the present research.
CS ChemDraw Ultra 7.01, Cambridge Soft Corp. (http://www.cambridgesoft.Com), USA.
Omega, OpenEye Scientific Software (www.eyesopen.com), USA.
Fred (Version 2.1.2), OpenEye Scientific Software (www.eyesopen.com), USA (Fred 2006).
Docking studies were performed using FRED and Omega software from OpenEye Scientific Inc (Santa Fe, NM, www.eyesopen.com) installed on Pentium 4 PC.
Preparation of DPP IV crystal structure
The 3D coordinates of DPP IV were retrieved from the Protein Data Bank (PDB code: 2G63)[Citation49]. The selected structure is of the best 3D resolution (2.0 å) compared to other available DPP IV structures. The co-crystallized ligand was extracted and hydrogen atoms were added to the protein utilizing Discovery Studio (DS) Visualizer 1.7 (Accelrys Inc., San Diego, California, www.accelrys.com). The protein structure was utilized in subsequent docking experiments without energy minimization. Explicit water molecules were kept in the structure.
Docking simulations
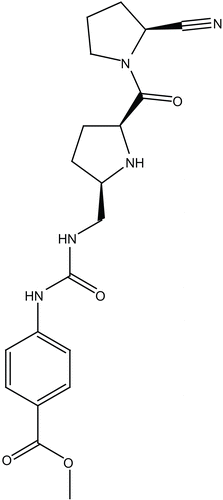
The chemical structure of berberine was sketched in Chemdraw Ultra (7.0), saved in MDL molfile format. The compound was docked into the binding site of DPP IV (PDB code: 2G63, resolution = 2.0 å) [Citation49] employing FRED software [Citation46]. This docking engine takes a multiconformer database of one or more ligands, a target protein structure, a box defining the active site of the protein based on the co-crystallized ligand and several optional parameters as input. The ligand conformers and protein structure are treated as rigid during the docking process. FRED’s docking strategy is to exhaustively score all possible positions of each ligand in the active site [Citation46]. The exhaustive search is based on rigid rotations and translations of each conformer. Therefore, it avoids sampling issues associated with stochastic methods; semi random method. The conformational space of berberine was explored using OMEGA software. The best docking conditions utilized were those succeeded to retrieve the pose of the co-crystallized ligand (cyanopyrrolidine inhibitor 24b [Citation49], ). In the current docking experiment the following docking parameters were employed
Addbox: Optional parameter that adjusts the geometry of the box definining the active site by extending each edge of the box by the specified number of Angstroms. (Type = float, default = 0). The value was set to 2.00.
Num_poses: this parameter specifies the number of poses to be returned by the exhaustive search. Poses will be the top scoring poses selected from the list of all poses and scored by the scoring functions specified by the -exhaustive scoring. Any number greater than 0 is a valid setting, although 10-1000 is a recommended reasonable range. (Type = integer, default = 100). The used value was 1000.
Num_alt_poses: This flag specifies how many alternate poses, in addition to the top consensus structure pose, will be passed out of the consensus structure step. Legal values are between 0 and 99.The value was set to be 10.
Clash_checking. This parameter specifies at what fraction of the sum of clash radii a ligand and protein heavy atom are considered to clash. This clash criterion is used to create the negative image of the receptor site. Allowable values are between 0.0 and 1, however values below 0.5 are not recommended. (Type = float, default 0.75). The value was set to be 0.5.
The docked poses were scored by the Chemgauss2 scoring function and the highest ranking poses were retained for evaluation.
Measurement of DPP IV inhibition
Berberine was dissolved in DMSO and diluted with the assay buffer (Tris, pH 7.5). The DMSO concentration was less than 1.0 % in all experiments and controls. The assays were conducted using DPP IV Drug Discovery Kit (Biomol, Germany), which is based on the cleavage of chromogenic substrate (H-Gly-Pro-para-Nitroaniline) by DPP IV to release para-nitroaniline (pNA) measured at 405 nm [Citation50]. Briefly, recombinant DPP IV was diluted in Tris buffer (pH 7.5, 50 mM) to a final enzymatic solution of 17.34μU/μL. Subsequently, appropriate volumes of the berberine stock solutions were added to a 15 μL aliquot of the enzymatic solution, then the volume completed to 50 μL with Tris buffer. The assay mixture was incubated at 37°C for 10 min. Finally, 50 μL of the substrate solution (0.20 mM in Tris buffer) was added and the absorbance was measured at 405 nm in a microplate reader (BioTek, USA) and the rate of reduction of substrate absorbance was evaluated over 20 min. and compared to a negative control (enzymatic solution without inhibition). A standard DPP IV Inhibitor (P32/98 from Biomol, Germany) was employed as a positive control [Citation51].
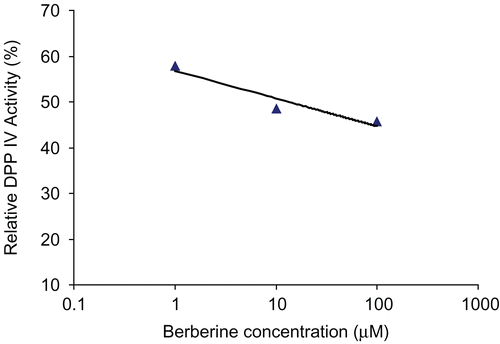
The percent activity was plotted against the logarithmic transformation of the corresponding berberine concentrations for determination of the IC50 value.
Results and discussion
Berberine is based on tetrahydroisoquinoline scaffold, which is an important structural motif commonly encountered in natural alkaloids of interesting biological profiles. In particular, this framework has become widely identified as “privileged” structure or pharmacophore that can bind to multiple target proteins [Citation52,Citation53]. This fact combined with the similarity between berberine and benzoquinolizine-based potent DPP IV inhibitors scaffold (e.g., compound ) [Citation38], prompted us to evaluate possible binding interaction(s) between berberine and DPP IV as an additional possible mechanism explaining its hypoglycemic action. Despite that the hypoglycemic action of berberine has been recently attributed to its potent anti-h-PTP1B properties [Citation37], its privileged structure suggests other possible hypoglycemic mechanisms [Citation52,Citation53].
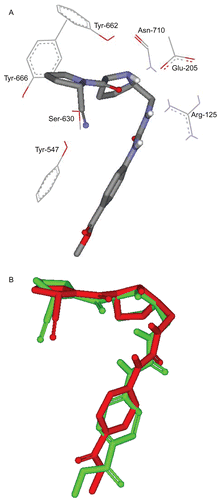
Our efforts commenced by evaluating the possibility of binding via computer-aided molecular modeling techniques. Accordingly, we docked berberine into the binding pocket of DPP IV (PDB code: 2G63). The binding site was defined from the crystallographic structure of a bound high-affinity ligand ( and , see Docking Simulations under Methods). Docking simulation suggested ten discrete binding modes for berberine within DPP IV. The molecular interactions of the highest ranking binding mode can be summarized in and . Clearly from the figures, the cationic nitrogen of berberine interacts with the negative charged carboxylate moiety of Glu-205, which is an important element of the anionic trap in DPP IV binding pocket [Citation54]. On the other hand, the dimethoxylated aromatic ring in berberine is situated at around 4.2 å from the guanidine moiety of Arg-125 suggesting a charge transfer attraction between the electron-rich aromatic ring and the positive charged guanidine group. Moreover, the oxygen of the dioxymethylene ring is hydrogen bonded to Ser-630 (1.8 å). Finally, the dioxymethylene substituted aromatic ring of berberine is well fitted in the hydrophobic S1 pocket; nearly 2.5 and 3.3 å away from the phenyl ring of Tyr-662 and Tyr-666 respectively, which suggests the existence of van der Waals’ aromatic stacking attraction between them. Overall, the three attractive interactions cooperate in stabilizing the proposed complex.
Figure 4. Perspective cartoon view of DPP IV co-crystallized with cyanopyrrolidine inhibitor. [PDB code: 2G63[49]]
![Figure 4. Perspective cartoon view of DPP IV co-crystallized with cyanopyrrolidine inhibitor. [PDB code: 2G63[49]]](/cms/asset/4bbf87ac-b29a-44ff-bfb9-b2d029c70161/ienz_a_361244_f0004_b.gif)
Figure 5. Perspective cartoon view of the highest ranking binding mode of berberine, as suggested by the Chemgauss2 scoring function, docked in DPP IV. [PDB code: 2G63[49]]
![Figure 5. Perspective cartoon view of the highest ranking binding mode of berberine, as suggested by the Chemgauss2 scoring function, docked in DPP IV. [PDB code: 2G63[49]]](/cms/asset/f0ca35a9-0903-4cb2-af57-0de53323ca92/ienz_a_361244_f0005_b.gif)
Figure 6. Detailed view of the docked berberine structure and the corresponding interacting amino-acid moieties within the binding site of DPP IV. [PDB code: 2G63[49]]
![Figure 6. Detailed view of the docked berberine structure and the corresponding interacting amino-acid moieties within the binding site of DPP IV. [PDB code: 2G63[49]]](/cms/asset/d6b5a19f-a267-45bd-900f-6ac3be3bf4a9/ienz_a_361244_f0006_b.gif)
However, to validate our docking-scoring procedure, we employed the same conditions to dock a well known DPP IV inhibitor (cyanopyrrolidine [Citation49], ) into the binding pocket of this enzyme. Our docking simulation resulted in a very close model to the crystallographic structure, which supports our conclusions regarding berberine/DPP IV binding (). Interestingly, similar interactions pattern has been observed for the co-crystallized compound ().
Figure 7. The highest ranking binding mode of the cyanopyrolidine inhibitor as suggested by the docking-scoring conditions. (A) Detailed view of the docked structure and the corresponding interacting amino-acids. (B) Comparison between the docked conformer/pose of inhibitor cyanopyrolidine (green) as produced by the docking simulation and the crystallographic structure of this inhibitor within DPP IV (red, PDB code: 2G63).
The proposed inhibitory action of berberine was experimentally validated against recombinant DPP IV using the H-Gly-Pro-pNA as substrate. The enzymatic reaction progression was monitored through the release of para-nitroaniline. The in vitro activity was expressed as the concentration of berberine that inhibited enzyme activity by 50% (IC50). The inhibitory action of berberine on DPP IV is estimated by an IC50 of 13.3 μM (). On the other hand, the IC50 of the positive control (P32/98) was estimated to be 131 nM. The DPP IV inhibitory activity could explain the anti-diabetic activity of berberine including the decrease of fasting blood glucose level, the increase in insulin secretion and the improvement in oral glucose tolerance tests (OGTT) observed in animal studies [Citation29].
Conclusion
The results of this study proved, through experimental testing and theoretical docking simulations, that berberine inhibits DPP IV, which could explain, at least partially, the reported use of berberine as an effective anti-hyperglycemic agent. The DPP IV inhibition activity along with the previously reported potent anti-h-PTP 1B action [Citation37] revealed a novel dual natural h-PTP 1B/DPP IV inhibitor. This duality is the first to be reported for such important, validated, anti-hyperglycemia targets.
Acknowledgements
This project was sponsored by the Deanship of Scientific Research at the University of Jordan. The authors wish to thank the Deanship of Scientific Research at the University of Jordan for providing funds. The authors would also like to thank the OpenEye Scientific Software for providing us with a free license for FRED software (FRED, version 2.1.2)
Declaration of interest: The authors report no conflicts of interest.
References
- Wild S, Roglic G, Green A, Sicree R, King H. Diabetes Care 2004; 27:1047–1053
- Saltiel AR. Cell 2001;104:517–529.
- Kreymann B, Williams G, Ghatei MA, Bloom SR. Lancet 1987;2:1300–1304.
- Dupre J, Ross SA, Watson D, Brown JC. J Clin Endocrinol Metab 1973;37:826.
- Nauck MA, Homberger E, Siegel EG, Allen RC, Eaton RP, Ebert R, Creutzfeldt W. JClin Endocrinol Metab 1986;63:492.
- Nauck MA, Baller BMeier JJ., Diabetes 2004;53:190–196.
- Drucker DJ. Diabetes Care 2003;26:2929–40.
- Lynn FC, Pamir N, Ng EH, McIntosh CH, Kieffer TJ, Pederson RA. Diabetes 2001;50:1004–11.
- Toft-Nielsen MB, Madsbad S, Holst JJ. J Clin Endocrinol Metab 2001;86:3853–60.
- Vilsboll T, Krarup T, Madsbad S, Holst JJ. Diabetologia 2002;45:1111–9.
- Deacon CF, Holst JJ. Int J Biochem Cell Biol2006;38:831–844
- Nauck MA, Wollschlager D, Werner J, Holst JJ, ørskov C, Creutzfeldt W, Willms B. Diabetologia1996;39:1546–1553.
- Salvatore T, Carbonara O, Cozzolino D, Torella R, Sasso FC. Curr Diabetes Rev 2007;3(1):15–23.
- Mentlein R, Gallwitz B, Schmidt WE. Eur J Biochem 1993;214:829–35.
- Ahren B, Landin-Olsson M, Jansson P, Svensson M, Holmes D, Schweizer A. J Clin Endocrinol Metab 2004;89:2078.
- Brubaker P, Drucker D. Endocrinology 2004;145:2653–2659.
- Meier JJ, Gallwitz B, Kask B, Deacon CF, Holst JJ, Schmidt WE, Nauck MA. Diabetes 2004;53: S220–4.
- Vilsboll T, Krarup T, Madsbad S, Holst JJ. Diabetic Med 2001;18(2):144–149.
- Weber AE. J Med Chem 2004;47:4135.
- Han Y, Lee JH. Biol Pharm Bull 2005;28:541–544.
- Freile ML, Giannini F, Pucci G, Sturniolo A, Rodero L, Pucci O, Balzareti V, Enriz RD. Fitoterapia 2003;74:702–705.
- Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K. Proc Natl Acad Sci USA 2000;97:1433–1437.
- Kuo CL, Chi CW, Liu TY. In Vivo 2005;19:247–252.
- Kuo CL, Chi CW, Liu TY. Cancer Lett 2004;203:127–137.
- Kong W, Wei J, Abidi P, Lin M, Inaba S, Li C, Wang Y, Wang Z, Si S, Pan H, Wang S, Wu J, Li Z, Liu J, Jiang JD. Nat Med 2004;10:1344–1351.
- Peng WH, Wu CR, Chen CS, Chen CF, Leu ZC, Hsieh MT. Life Sci 2004;75:2451–2462.
- Hong Y, Hui SS, Chan BT, Hou J. Life Sci 2003;72:2499–2507.
- Lin S, Tsai SC, Lee CC, Wang BW, Liou JY, Shyu KG. Mol Pharmacol 2004;66:612–619.
- Leng San-hua, Lu F, Xu Li-jun. Acta Pharmacol Sin 2004;25:496–502.
- Yin J, Hu R, Chen M, Tang J, Li F, Yang Y, Chen J. Metab Clin Exp 2002;51:1439–1443.
- Gao CR, Zhang JQ, Huang QL. Chinese J Inetegr Trad Western Med 1997;17:162–164.
- Chen QM, Xie MZ. Yaoxue Xuebao 1987;22:161–165.
- Jun Y, Huili X, Jianping Ye. Metabolism 2008;57:712–717
- Ni YX. Chinese journal of modern developments in traditional medicine (Zhong xi yi jie he za zhi). 1988;8:711–713.
- Wei J, Jiang J, Wang Z, Pan H. Application of berberine as insulin sensitizer. Faming Zhuanli Shenqing Gongkai Shuomingshu. 2003, 20030129, p 6.
- Pan GY, Huang ZJ, Wang GJ, Fawcett JP, Liu XD, Zhao XC, Sun JG, Xie YY. Planta Med 2003;69:632–636.
- Bustanji Y, Taha MO, Yousef A, Al-Bakri, AG. J Enz Inhib Med Chem 2006; 21(2):163–171.
- Luebbers T, Boehringer M, Gobbi L, Hennig M, Hunziker D, Kuhn B, Loeffler B, Mattei P, Narquizian R, Peters J, Ruff Y, Wessel HP, Wyss P. Bioorg Med Chem Lett 2007;17(11):2966–2970.
- Mestres J, Knegtel RMA. Perspect Drug Discov Des 2000;20:191–207.
- Monard G, Merz KM. Accounts Chem Res 1999;32:904–911.
- Morris GM, Olson AJ, Goodsell DS. Methods Princ Med Chem 2000;8:31–48.
- Vieth M, Hirst JD, Dominy BN, Daigler H, Brooks CL. J Comput Chem 1998;19:1623–1631.
- Gilson MK, Given JA, Bush BL, McCammon JA. Biophys J 1997;72:1047–1069.
- Kontoyianni M, McClellan LM, Sokol GS. J Med Chem 2004;47:558–565.
- Bissantz C, Folkers G, Rognan D. J Med Chem 2000;43:4759–4767.
- Fred, 2006. version 2.1.2 Users’ Manual, OpenEye Scientific Software Inc., Santa Fe, New Mexico.
- McGaughey GB, Sheridan RP, Bayly CI, Culberson JC, Kreatsoulas C, Lindsley S, Maiorov V, Truchon J, Cornell WD. J Chem Inf Model 2007;47(4):1504–1519.
- Ragno R, Mai A, Simeoni S, Caroli A, Caffarelli E, La Neve P, Gioia U, Bozzoni I. Discovery of XendoU Inhibitors through Multi-Docking Virtual Screening. Frontiers in CNS and Oncology Medicinal Chemistry, ACS-EFMC, 2007; 7–9.
- Pei Z, Li X, Longenecker K, Von Geldern TW, Wiedeman PE, Lubben TH, Zinker BA, Stewart K, Ballaron SJ, Stashko MA, Mika AK, Beno DW, Long M, Wells H., Kempf-Grote AJ, Madar DJ, McDermott TS, Bhagavatula L, Fickes MG, Pireh D, Solomon LR, Lake MR, Edalji R, Fry EH, Sham HL, Trevillyan JM. J Med Chem. 2006;49: 3520–3535
- Al-masri I, Mohammad M, Taha M. ChemMedChem, In press
- Pospisilik A, Martin J, Doty T, Ehses A, Pamir N, Lynn C, Piteau S, Demuth H-U, McIntosh S, Pederson A. Diabetes 2003;52(3):741–750.
- Scott JD, Williams RM. Chem Rev 2002;102:1669–1730.
- Yang J, In J, Lee M, Kwak J, Lee H, Lee S, Kang H, Suh Y, Jung JS. Bull Korean Chem Soc 2007;28(8):1401–1404.
- Kuhn B, Hennig M, Mattei P. Curr Top Med Chem 2007;7:609–619