Abstract
ARS-interacting multifunctional proteins 2 (AIMP2) is known to be a powerful tumour suppressor. However, the target AIMP2-DX2, AIMP2-lacking exon 2, is often detected in many cancer patients and cells. The predominant approach for targeting AIMP-DX2 has been attempted via small molecule mediated inhibition, but due to the lack of satisfactory activity against AIMP2-DX2, new therapeutic strategies are needed to develop a novel drug for AIMP2-DX2. Here, we report the use of the PROTAC strategy that combines small-molecule AIMP2-DX2 inhibitors with selective E3-ligase ligands with optimised linkers. Consequently, candidate compound 45 was found to be a degrader of AIMP2-DX2. Together, these findings demonstrate that our PROTAC technology targeting AIMP2-DX2 would be a potential new strategy for future lung cancer treatment.
Keywords:
Introduction
Multi-tRNA synthetase complex (MSC) plays an essential role in charging of tRNA with the cognate amino acid for synthesising peptide. It consists of nine types of Aminoacyl-tRNA Synthetases (ARS) and three types of ARS-interacting multifunctional proteins (AIMPs). Among the three AIMPs, AIMP2, which consists of four exons, is known as a powerful tumour suppressor by mediating various transduction systemsCitation1–4. A splicing variant of AIMP2, AIMP2 lacking exon 2 (AIMP2-DX2), is established as an oncogene and is highly expressed in lung cancer cells and lung cancer patient tissuesCitation5–7. This is because AIMP2-DX2 binds to its binding partners FBP, TRAF2, and p53 and competitively inhibits the anticancer function of AIMP2 to induce cancer and also stabilises the cancer driver gene KRASCitation5,Citation7,Citation8. Recently, Lim et al.Citation9 discovered that the level of AIMP2-DX2 in cells can be regulated by Siah1 protein, a specific E3 ligase of AIMP2-DX2, via an ubiquitination-mediated degradation sequences. However, when 70 kDa heat shock proteins (HSP70s) selectively bind to AIMP2-DX2, it blocks the binding of AIMP2-DX2 and Siah1 protein, resulting in an increase in AIMP2-DX2 levelCitation9. Although many studies have been conducted so far to identify small molecules targeting AIMP2-DX2 itself using conventional methods, the development of new lung cancer drug that is fine-tuned to chemically inhibit the interaction of HSP70 and AIMP2-DX2 and increase the accessibility of Siah1 and AIMP2-DX2 has not been attemptedCitation4,Citation9–13. To the best of our knowledge, a comprehensive study of E3 ligase ligand and linker length activity relationships with a PROTAC targeting AIMP-DX2 has not been forthcoming in the literature. Accordingly, this study presents the PROTAC technique as a promising alternative approach against AIMP2-DX2 targeting lung cancer.
Proteolysis targeting chimaera (PROTAC) technology has received great attention in drug discovery and development areas over the past few years as the clinical results of anticancer drugs applied with PROTAC technology were positively announced. PROTAC is a heterobifunctional molecules and consists of three parts: target ligand, linker, and E3 ligand. It is designed to bind with the target protein and E3 ligase (). This new platform technology follows a well-known ubiquitin-proteasome system. PROTAC can recruit E3 ubiquitin ligase to a target protein of interest, resulting in polyubiquitination of the protein of interest which is subsequently degraded via 26S proteasomeCitation14. In addition, PROTAC can act as a catalyst, making it reusable. It can exert the desired pharmacological effect at a much lower concentration compared to conventional small molecule inhibitors, thereby reducing the toxicity problem of small molecule inhibitorsCitation15.
In this study, we describe our efforts in the discovery of potent AIMP2-DX2 degraders utilising CRBN E3 ligases and various linkers, which resulted in a promising compound with anticancer activities. These data demonstrate the importance and potential for developing PROTAC-based therapeutic molecules for the AIMP2-DX2 targeted lung cancer drug.
Experimental section
General chemistry
All commercially available chemicals were of reagent grade and were used without further purification. Solvents were dried with standard procedures. All of the reactions were purified by flash column chromatography using CombiFlash® Rf 200 system (Teledyne Isco., Lincoln, NE). Additionally, thin-layer chromatography was performed on 0.25 mm silica gel 60-F254 plates (Merck, Kenilworth, NJ). Visualisation was accomplished with 254 nm of UV light or potassium permanganate staining followed by heating. The proton nuclear magnetic resonance (1H-NMR) spectra were determined on a Varian (400 and 100 MHz) or Bruker (400 and 100 MHz). 1H NMR (at 400 MHz) was reported on a Varian 400 MHz spectrometer and are reported in ppm using solvent as an internal standard (DMSO-d6 at 2.50, Acetone-d6 at 2.05, at 3.31 for 1H NMR). Data reported as (s = singlet, d = doublet, t = triplet, q = quartette, m = multiplet, br = broad; coupling constant(s) in Hz; integration). LC/MS data were obtained using a Waters 2695-ZQ2000 system and Waters ACQUITY UPLC-SQ Detector 2.
Specific experimental procedures and product characterisation
General procedure for the click reaction
(+)-Sodium L-ascorbate (0.15 equiv.) and copper (II) sulphate pentahydrate (0.05 equiv.) were added to a stirred solution of azide (1equiv.) and alkyne (1 equiv.) in DMSO at 25 °C. The mixture was stirred at 25 °C for overnight before being quenched with aq. NaHCO3 and extracted with dichloromethane. The combined organic phases were washed with water and brine then dried (Na2SO4), filtered, and concentrated under reduced pressure. The residue thus obtained was subjected to column chromatography to give the desired product.
(2S)-2-((4-((4-((2–(2-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)oxy)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (13)
The title compound was prepared from azide 11 and alkyne 12a according to general procedure as a light green solid (14.0 mg, 27%). 1H NMR (400 MHz, Acetone-d6) δ 9.93 (s, 1H), 9.90 (s, 1H), 8.96 (s, 1H), 7.84 (s, 1H), 7.77 (t, J = 8.8 Hz, 1H), 7.58 (d, J = 8.0 Hz, 2H), 7.49–7.44 (m, 3H), 7.42–7.30 (m, 3H), 7.14 (d, J = 8.0 Hz, 2H), 7.10–7.06 (m, 2H), 6.95 (t, J = 7.4 Hz, 1H), 6.88 (t, J = 9.2 Hz, 2H), 6.81–6.88 (m, 1H), 5.55 (s, 2H), 5.11 (dd, J = 5.2 and 12.4 Hz, 1H), 4.60 (s, 2H), 4.34 (t, J = 4.8 Hz, 2H), 4.17 (brs, 1H), 3.83 (t, J = 4.8 Hz, 2H), 3.77–3.75 (m, 4H), 3.73–3.71 (m, 2H), 3.66–3.63 (m, 2H), 3.26 (dd, J = 5.2 and 12.4 Hz, 1H), 3.10–3.05 (m, 4H), 3.03–2.71 (m, 4H), 2.22–2.10 (m, 1H); LCMS (ESI) m/z 960 [M + H]+.
(2S)-2-((4-((4-((2–(2–(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)oxy)ethoxy)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (14)
The title compound was prepared from azide 11 and alkyne 12b according to general procedure as a pale yellow solid (3.0 mg, 6%). 1H NMR (400 MHz, Acetone-d6) δ 9.93 (s, 1H), 9.91 (s, 1H), 8.95 (s, 1H), 7.83 (s, 1H), 7.77 (t, J = 8.4 Hz, 1H), 7.60 (d, J = 7.6 Hz, 2H), 7.48–7.42 (m, 3H), 7.43–7.30 (m, 3H), 7.15 (d, J = 8.0 Hz, 2H), 7.10–7.06 (m, 2H), 6.95 (t, J = 7.6 Hz, 1H), 6.89–6.86 (m, 2H), 6.80 (d, J = 8.4 Hz, 1H), 5.55 (s, 2H), 5.10 (dd, J = 5.2 and 12.8 Hz, 1H), 4.59 (s, 2H), 4.35 (t, J = 4.8 Hz, 2H), 4.17–4.16 (m, 1H), 3.86 (t, J = 4.8 Hz, 2H), 3.76 (t, J = 4.8 Hz, 4H), 3.69–3.67 (m, 2H), 3.62–3.55 (m, 6H), 3.24 (dd, J = 6.0 and 14.8 Hz, 1H), 3.08 (t, J = 4.8 Hz, 4H), 3.03–2.74 (m, 4H), 2.22–2.17 (m, 1H); LCMS (ESI) m/z 1002 [M-H]-.
(2S)-2-((4-((4–(13-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)oxy)-2,5,8,11-tetraoxatridecyl)-1H-1,2,3-triazol-1-yl)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (15)
The title compound was prepared from azide 11 and alkyne 12c according to general procedure as a pale-yellow solid (2.0 mg, 4%). 1H NMR (400 MHz, Acetone-d6) δ 9.91 (brs, 2H), 8.96 (brs, 1H), 7.84 (s, 1H), 7.79 (t, J = 8.4 Hz, 1H), 7.60 (d, J = 8.4 Hz, 2H), 7.49–7.42 (m, 3H), 7.34–7.30 (m, 3H), 7.15 (d, J = 8.0 Hz, 2H), 7.08 (dd, J = 8.4 and 6.4 Hz, 2H), 6.95 (dd, J = 8.0 and 6.8 Hz, 1H), 6.88 (d, J = 8.8 Hz, 2H), 6.79 (brs, 1H), 5.56 (s, 2H), 5.10 (dd, J = 5.6 and 12.4 Hz, 1H), 4.59 (s, 2H), 4.36 (t, J = 4.8 Hz, 2H), 4.16 (brs, 1H), 3.86 (t, J = 4.8 Hz, 2H), 3.76 (t, J = 4.8 Hz, 4H), 3.70–3.67 (m, 2H), 3.61–3.51 (m, 10H), 3.24 (dd, J = 6.2 and 14.6 Hz, 1H), 3.08 (t, J = 4.8 Hz, 4H), 3.05–2.74 (m, 4H), 2.21–2.10 (m, 1H); LCMS (ESI) m/z 1046 [M-H]-.
(2S)-2-((4-((4–(16-((2–(2,6-dioxopiperidin-3-yl)-1,3-Dioxoisoindolin-4-yl)oxy)-2,5,8,11,14-pentaoxahexadecyl)-1H-1,2,3-triazol-1-yl)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (16)
The title compound was prepared from azide 11 and alkyne 12d according to general procedure as a pale-yellow solid (22.0 mg, 37%). 1H NMR (400 MHz, Acetone-d6) δ 9.92 (brs, 2H), 8.98 (brs, 1H), 7.85 (s, 1H), 7.79 (t, J = 8.8 Hz, 1H), 7.61 (d, J = 8.4 Hz, 2H), 7.50–7.42 (m, 3H), 7.34–7.31 (m, 3H), 7.16 (d, J = 8.4 Hz, 2H), 7.08 (t, J = 7.2 Hz, 2H), 6.95 (t, J = 7.2 Hz, 1H), 6.87 (d, J = 12.4 Hz, 2H), 6.83 (brs, 1H), 5.56 (s, 2H), 5.10 (dd, J = 5.2 and 12.8 Hz, 1H), 4.59 (s, 2H), 4.37 (t, J = 4.8 Hz, 2H), 4.16 (t, J = 7.6 Hz, 1H), 3.87 (dd, J = 4.8 Hz, 2H), 3.77–3.75 (m, 4H), 3.70–3.68 (m, 2H), 3.62–3.51 (m, 14H), 3.25 (dd, J = 5.8 and 14.6 Hz, 1H), 3.10–3.07 (m, 4H), 3.05–2.73 (m, 4H), 2.23–2.19 (m, 1H); LCMS (ESI) m/z 1090 [M-H]-.
(2S)-2-((4-((4-((3-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3-oxopropoxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (18)
The title compound was prepared from azide 11 and alkyne 17a according to general procedure as a pale yellow solid (15.8 mg, 31%); 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s, 1H), 10.71 (s, 1H), 9.86 (s, 1H), 9.67 (s, 1H), 8.51 (d, J = 8.4 Hz, 1H), 8.16 (brs, 1H), 8.03 (s, 1H), 7.82 (t, J = 7.6 Hz, 1H), 7.61–7.56 (m, 3H), 7.41 (d, J = 7.6 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.19–7.14 (m, 4H), 7.06–7.01 (m, 2H), 6.91 (t, J = 7.6 Hz, 1H), 6.84 (d, J = 8.8 Hz, 2H), 5.53 (s, 2H), 5.13 (dd, J = 5.6 and 12.8 Hz, 1H), 4.60 (s, 2H), 4.09 (t, J = 6.8 Hz, 1H), 3.78–3.71 (m, 6H), 3.07–3.03 (m, 5H), 2.88–2.83 (m, 2H), 2.72 (t, J = 6.0 Hz, 2H), 2.63–2.53 (m, 2H), 2.07–2.04 (m, 1H); LCMS (ESI) m/z 943 [M + H]+.
(2S)-2-((4-((4-((2–(3-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3-oxopropoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (19)
The title compound was prepared from azide 11 and alkyne 17b according to general procedure as a pale yellow solid (23.0 mg, 43%); 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s, 1H), 10.71 (s, 1H), 9.86 (s, 1H), 9.67 (s, 1H), 8.51 (d, J = 8.4 Hz, 1H), 8.16 (brs, 1H), 7.99 (s, 1H), 7.81 (t, J = 7.6 Hz, 1H), 7.59 (t, J = 8.4, 3H), 7.41 (d, J = 7.6 Hz, 1H), 7.29 (d, J = 8.0 Hz, 1H), 7.19–7.14 (m, 4H), 7.06–7.02 (m, 2H), 6.91 (t, J = 7.6 Hz, 1H), 6.84 (d, J = 9.2 Hz, 2H), 5.52 (s, 2H), 5.13 (dd, J = 5.6 and 12.8 Hz, 1H), 4.51 (s, 2H), 4.09 (t, J = 7.2 Hz, 1H), 3.73–3.71 (m, 6H), 3.59 (s, 4H), 3.07–3.03 (m, 5H), 2.94–2.83 (m, 2H), 2.69 (t, J = 6.0 Hz, 2H), 2.63–2.55 (m, 2H), 2.09–2.05 (m, 1H); LCMS (ESI) m/z 987 [M + H]+.
(2S)-2-((4-((4-((2–(2-(3-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3-oxopropoxy)ethoxy)ethoxy)methyl)-1H-1,2,3-triazol-1-yl)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (20)
The title compound was prepared from azide 11 and alkyne 17c according to general procedure as a yellow solid (15.5 mg, 28%); 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s, 1H), 10.71 (s, 1H), 9.85 (s, 1H), 9.66 (s, 1H), 8.53 (d, J = 8.4 Hz, 1H), 8.15 (d, J = 8.8 Hz, 1H), 8.00 (s, 1H), 7.81 (t, J = 7.6 Hz, 1H), 7.58 (t, J = 8.0 Hz, 3H), 7.40 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.19–7.14 (m, 4H), 7.05–7.01 (m, 2H), 6.91 (t, J = 7.2 Hz, 1H), 6.83 (d, J = 8.8 Hz, 2H), 5.53 (s, 2H), 5.13 (dd, J = 5.6 and 12.8 Hz, 1H), 4.48 (s, 2H), 3.72–3.71 (m, 6H), 3.55–3.52 (m, 9H), 3.07–3.02 (5H), 2.93–2.82 (m, 2H), 2.68 (t, J = 6.0 Hz, 2H), 2.58–2.54 (m, 2H), 2.07–2.05 (m, 1H); LCMS (ESI) m/z 1029 [M-H]-.
1–(1-(4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)benzyl)-1H-1,2,3-triazol-4-yl)-N-(2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)-2,5,8,11-tetraoxatetradecan-14-amide (21)
The title compound was prepared from azide 11 and alkyne 17d according to general procedure as a brown solid (4.5 mg, 9%); 1H NMR (400 MHz, DMSO-d6) δ 11.11 (s,1H), 10.71 (s, 1H), 9.85 (s, 1H), 9.66 (s, 1H), 8.54 (d, J = 8.4 Hz, 1H), 8.15 (brs, 1H), 8.01 (s, 1H), 7.84 (t, J = 7.6 Hz, 1H), 7.62–7.56 (m, 3H), 7.40 (d, J = 8.0 Hz, 2H), 7.28 (d, J = 8.0 Hz, 1H), 7.19–7.14 (m, 4H), 7.05–7.01 (m, 2H), 6.91 (t, J = 7.2 Hz, 1H), 6.83 (d, J = 8.8 Hz, 2H), 5.53 (s, 2H), 5.13 (dd, J = 5.6 and 12.8 Hz, 1H), 4.50 (s, 2H). 4.09 (brs, 1H), 3.72–3.71 (m, 6H), 3.52–3.41 (m, 11H), 3.04–3.02 (m, 5H), 2.93–2.82 (m, 2H), 2.70–2.67 (m, 2H), 2.58–2.50 (m, 2H), 2.07–2.05 (m, 1H); LCMS (ESI) m/z 1073 [M-H]-.
1–(1-(4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)benzyl)-1H-1,2,3-triazol-4-yl)-N-(2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)-2,5,8,11,14-pentaoxaheptadecan-17-amide (22)
The title compound was prepared from azide 11 and alkyne 17e according to general procedure as a light brown solid (12.5 mg, 25%); 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s, 1H), 10.71 (s, 1H), 9.86 (s, 1H), 9.67 (s, 1H), 8.54 (d, J = 8.4 Hz, 1H), 8.16 (brs, 1H), 8.02 (s, 1H), 7.82 (t, J = 7.6, 1H), 7.59 (dd, J = 8.0 and 9.6 Hz, 3H), 7.41 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 7.6 Hz, 1H), 7.19–7.15 (m, 4H), 7.06–7.02 (m, 2H), 6.91 (t, J = 7.2 Hz, 1H), 6.84 (d, J = 8.4 Hz, 2H), 5.54 (s, 2H), 5.14 (dd, J = 4.4 and 12.4 Hz, 1H), 4.52 (s, 2H), 4.09 (brs, 1H), 3.73 (brs, 6H), 3.55–3.45 (m, 16H), 3.04 (m, 5H), 2.94–2.83 (m, 2H), 2.69 (brs, 2H), 2.64–2.55 (m, 2H), 2.09–2.07 (m, 1H); LCMS (ESI) m/z 1117 [M-H]-.
1–(1-(4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)benzyl)-1H-1,2,3-triazol-4-yl)-N-(2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)-2,5,8,11,14,17-hexaoxaicosan-20-amide (23)
The title compound was prepared from azide 11 and alkyne 17f according to general procedure as a yellow solid (9.2 mg, 22%); 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s,1H), 10.71 (s, 1H), 9.86 (s, 1H), 9.67 (s, 1H), 8.54 (d, J = 8.4 Hz, 1H), 8.16 (brs, 1H), 8.02 (s, 1H), 7.83 (dd, J = 7.2 and 8.0 Hz, 1H), 7.61–7.57 (m, 3H), 7.41 (d, J = 7.6 Hz, 1H), 7.29 (d, J = 8.0 Hz, 1H), 7.17(dd, J = 9.2 and 8.4 Hz, 4H), 7.06–7.02 (m, 2H), 6.91 (t, J = 7.6 Hz, 1H), 6.84 (d, J = 8.4 Hz, 1H), 5.54 (s, 2H), 5.13 (dd, J = 5.2 and 12.4 Hz, 1H), 4.52 (s, 2H), 4.10–4.04 (m, 1H), 3.73 (brs, 6H), 3.56–3.45 (m, 20H), 3.04 (brs, 5H), 2.94–2.82 (m, 2H), 2.69–2.68 (m, 2H), 2.64–2.51 (m, 2H), 2.09–2.06 (m, 1H); LCMS (ESI) m/z 1161 [M-H]-.
General procedure for amide coupling
To a solution of acid (1 equiv.), amine (1.2 equiv.), HATU (1.2 equiv.) in DMF, DIPEA (1.2 equiv.) was added. The mixture was stirred at 25 °C for overnight. The reaction mixture was quenched with water and extracted with ethyl acetate. The combined organic phases were washed with brine and dried (Na2SO4), filtered, and concentrated under reduced pressure. The residue thus obtained subjected to column chromatography to give the desired product.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(4–(2-((2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)oxy)acetamido)butyl)benzamide (25)
The title compound was prepared from 24 according to general procedure as a yellow solid (10.0 mg, 12%); 1H NMR (400 MHz, DMSO-d6) δ 11.09 (brs, 1H), 10.74 (s, 1H), 9.71 (s, 1H), 8.48 (t, J = 5.6 Hz, 1H), 8.31 (brs, 1H), 7.98 (t, J = 5.6 Hz, 1H), 7.81–7.76 (m, 3H), 7.68 (d, J = 8.4 Hz, 2H), 7.48 (d, J = 7.2 Hz, 1H), 7.40 (t, J = 8.4 Hz, 2H), 7.25 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 8.8 Hz, 2H), 7.07 (d, J = 2.0 Hz, 1H), 7.01 (t, J = 7.0 Hz, 1H), 6.91 (t, J = 7.0 Hz, 1H), 6.79 (d, J = 8.0 Hz, 2H), 5.11 (dd, J = 5.2 and 12.8 Hz, 1H), 4.77 (s, 2H), 4.14 (t, J = 7.2 Hz, 1H), 3.71 (t, J = 5.2 Hz, 4H), 3.27–3.19 (m, 4H), 3.09–2.99 (m, 5H), 2.89–2.84 (m, 2H), 2.67–2.57 (m, 2H), 2.04–2.01 (m, 1H), 1.53–1.51 (m, 4H); LCMS (ESI) m/z 933 [M + H]+.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(2–(2–(2–(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)oxy)acetamido)ethoxy)ethoxy)ethyl)benzamide (26)
The title compound was prepared from 24 according to general procedure as a yellow solid (30 mg, 39%); 1H NMR (400 MHz, DMSO-d6) δ 11.10 (s, 1H), 10.72 (s, 1H), 9.72 (s, 1H), 8.51 (t, J = 5.4 Hz, 1H), 8.30 (d, J = 8.4 Hz, 1H), 7.99 (t, J = 5.4 Hz, 1H), 7.81–7.73 (m, 3H), 7.66 (d, J = 8.4 Hz, 2H), 7.48 (d, J = 7.2 Hz, 1H), 7.39 (t, J = 8.8 Hz, 2H), 7.25 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 8.8 Hz, 2H), 7.07 (d, J = 2.0 Hz, 1H), 7.01 (t, J = 7.4 Hz, 1H), 6.90 (t, J = 7.2 Hz, 1H), 6.79 (d, J = 8.8 Hz, 2H), 5.10 (dd, J = 5.6 and 12.8 Hz, 1H), 4.77 (s, 2H), 4.16–4.10 (m, 1H), 3.71–3.69 (m, 4H), 3.55–3.45 (m, 8H), 3.33–3.30 (m, 2H), 3.01–2.88 (m, 5H), 2.86–2.82 (m, 2H), 2.61–2.54 (m, 2H), 2.05–1.98 (m, 1H), 1.23 (brs, 2H); LCMS (ESI) m/z 993 [M + H]+.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(2–(2-((2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-2-oxoethoxy)ethyl)benzamide (28)
The title compound was prepared from acid 24 and amine 27a according to general procedure as a yellow solid (27.4 mg, 55%); 1H NMR (400 MHz, DMSO-d6) δ 11.10 (s, 1H), 10.71 (s, 1H), 10.36 (s, 1H), 9.69 (s, 1H), 8.70 (d, J = 8.4 Hz, 1H), 8.58 (t, J = 5.6 Hz, 1H), 8.28 (brs, 1H), 7.86 (t, J = 7.6 Hz Hz, 1H), 7.75 (d, J = 8.4 Hz, 2H), 7.67–7.61 (m, 3H), 7.40 (d, J = 7.6 Hz, 1H), 7.24 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 8.8 Hz, 2H), 7.06 (d, J = 2.0 Hz, 1H), 7.00 (t, J = 7.2 Hz, 1H), 6.90 (t, J = 7.2 Hz, 1H), 6.79 (d, J = 9.2 Hz, 2H), 5.11 (dd, J = 5.6 and 12.8 Hz, 1H), 4.24 (s, 2H), 4.14–4.09 (m, 1H), 3.78 (t, J = 5.6 Hz, 2H), 3.70 (d, J = 4.4 and 4.8 Hz, 4H), 3.58 (dd, J = 5.6 and 11.6 Hz, 2H), 3.16 (d, J = 5.2 Hz, 1H), 3.08–2.99 (m, 5H), 2.90–2.81 (m, 2H), 2.70–2.56 (m, 1H), 2.03–2.00 (m, 1H); LCMS (ESI) m/z 905 [M + H]+.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(2–(2–(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-2-oxoethoxy)ethoxy)ethyl)benzamide (29)
The title compound was prepared from acid 24 and amine 27b according to general procedure as a yellow solid (24.2 mg, 47%); 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s, 1H), 10.70 (s, 1H), 10.35 (s, 1H), 9.70 (s, 1H), 8.69 (d, J = 8.4 Hz, 1H), 8.48 (t, J = 5.6 Hz, 1H), 8.28 (brs, 1H), 7.84 (t, J = 7.6 Hz, 1H), 7.72 (d, J = 8.8 Hz, 2H), 7.65–7.59 (m, 3H), 7.39 (d, J = 8.0 Hz, 1H), 7.24 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 8.8 Hz, 2H), 7.06 (d, J = 2.4 Hz, 1H), 7.00 (t, J = 7.2 Hz, 1H), 6.90 (t, J = 7.2 Hz, 1H), 6.78 (d, J = 9.2 Hz, 2H), 5.15 (dd, J = 5.6 and 12.8 Hz, 1H), 4.20 (s, 2H), 4.14–4.09 (m, 2H), 3.79–3.77 (m, 2H), 3.71–3.69 (m, 5H), 3.58 (t, J = 6.0 Hz, 2H), 3.17 (d, J = 5.2 Hz, 2H), 3.08–2.99 (m, 5H), 2.91–2.82 (m, 2H), 2.67–2.54 (m, 2H), 2.08–2.04 (m, 1H); LCMS (ESI) m/z 949 [M + H]+.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(2–(2–(2–(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-2-oxoethoxy)ethoxy)ethoxy)ethyl)benzamide (30)
The title compound was prepared from acid 24 and amine 27c according to general procedure as a yellow solid (17.2 mg, 32%); 1H NMR (400 MHz, DMSO-d6) δ 11.13 (s, 1H), 10.71 (s, 1H), 10.33 (s, 1H), 9.70 (s, 1H), 8.70 (d, J = 8.4 Hz, 1H), 8.47 (t, J = 5.6 Hz, 1H), 8.28 (brs, 1H), 7.84 (t, J = 7.6 Hz, 1H), 7.73 (d, J = 8.4 Hz, 2H), 7.66–7.60 (m, 3H), 7.40 (d, J = 8.0 Hz, 1H), 7.24 (d, J = 8.4 Hz, 1H), 7.17 (d, J = 9.2 Hz, 2H), 7.06 (d, J = 2.0 Hz, 1H), 7.00 (t, J = 7.2 Hz, 1H), 6.90 (t, J = 7.2 Hz, 1H), 6.79 (d, J = 9.2 Hz, 2H), 5.14 (dd, J = 5.6 and 12.8 Hz, 1H), 4.18 (s, 2H), 4.14–4.09 (m, 2H), 3.75–3.69 (m, 6H), 3.67–3.65 (m, 2H), 3.56–3.50 (m, 3H), 3.17 (d, J = 5.2 Hz, 4H), 3.08–2.99 (m, 5H), 2.93–2.83 (m, 2H), 2.67–2.54 (m, 2H), 2.10–2.05 (m, 1H); LCMS (ESI) m/z 993 [M + H]+.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(17-((2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-17-oxo-3,6,9,12,15-pentaoxaheptadecyl)benzamide (31)
The title compound was prepared from acid 24 and amine 27d according to general procedure as a yellow solid (28.1 mg, 48%); 1H NMR (400 MHz, DMSO-d6) δ 11.13 (s, 1H), 10.73 (s, 1H), 10.35 (s, 1H), 9.72 (s, 1H), 8.72 (d, J = 8.8 Hz, 1H), 8.51 (t, J = 5.6 Hz, 1H), 8.32 (brs, 1H), 7.85 (t, J = 7.6 Hz, 1H), 7.75 (d, J = 8.4 Hz, 2H), 7.67–7.61 (m, 3H), 7.41 (d, J = 7.6 Hz, 1H), 7.25 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 9.8 Hz, 2H), 7.06 (d, J = 2.0 Hz, 1H), 7.00 (t, J = 7.2 Hz, 1H), 6.90 (t, J = 7.2 Hz, 1H), 6.79 (d, J = 9.2 Hz, 2H), 5.16 (dd, J = 5.6 and 13.2 Hz, 1H), 4.20 (s, 2H), 4.13 (brs, 1H), 3.76–3.70 (m, 6H), 3.66–3.64 (m, 2H), 3.53–3.40 (m, 14H), 3.08–2.99 (m, 5H), 2.94–2.83 (m, 2H), 2.67–2.54 (m, 4H), 2.09–2.06 (m, 1H); LCMS (ESI) m/z [M + H]+.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(2–(2–(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethyl)benzamide (33)
The title compound was prepared from acid 24 and amine 33 according to general procedure as a yellow solid (10.7 mg, 21%); 1H NMR (400 MHz, DMSO-d6) δ 11.08 (s, 1H), 10.73 (s, 1H), 9.73 (s, 1H), 8.51 (t, J = 5.6 Hz, 1H), 8.32 (s, 1H), 7.75 (d, J = 8.4 Hz, 2H), 7.66 (d, J = 8.4 Hz, 2H), 7.56 (t, J = 8.0 Hz, 1H), 7.40 (d, J = 8.0 Hz, 1H), 7.25 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 8.8 Hz, 2H), 7.12–7.07 (m, 2H), 7.04–6.99 (m, 2H), 6.91 (t, J = 7.6 Hz, 1H), 6.80 (d, J = 9.2 Hz, 2H), 6.60 (t, J = 5.6 Hz, 1H), 5.05 (dd, J = 5.2 and 12.4 Hz, 1H), 4.13 (s, 1H), 3.72–3.70 (m, 4H), 3.62–3.53 (m, 8H), 3.46–3.41 (m, 4H), 3.08–2.99 (m, 5H), 2.91–2.82 (m, 2H), 2.67–2.54 (m, 2H) 2.02–1.94 (m, 1H); LCMS (ESI) m/z 935 [M + H]+.
4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)-N-(2–(2–(2–(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)ethyl)benzamide (34)
The title compound was prepared from acid 24 and amine 34 according to general procedure as a yellow solid (12.6 mg, 16%); 1H NMR (400 MHz, DMSO-d6) δ 11.08 (s,1H), 10.73 (s, 1H), 9.73 (s, 1H), 8.51 (t, J = 5.6 Hz, 1H), 8.33 (brs, 1H), 7.75 (d, J = 8.4 Hz, 2H), 7.66 (d, J = 8.4 Hz, 2H), 7.56 (t, J = 8.0 Hz, 1H), 7.40 (d, J = 8.0 Hz, 1H), 7.25 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 9.2 Hz, 2H), 7.12 (d, J = 8.8 Hz, 1H), 7.07 (d, J = 2.0 Hz, 1H), 7.04–6.99 (m, 2H), 6.90 (t, J = 7.6 Hz, 1H), 6.79 (d, J = 8.8 Hz, 2H), 6.57 (t, J = 5.6 Hz, 1H), 5.04 (dd, J = 5.6 and 12.8 Hz, 1H), 4.12 (t, J = 7.2 Hz, 1H), 3.72–3.69 (m, 4H), 3.59 (t, J = 5.6 Hz, 2H), 3.62–3.53 (m, 11H), 3.08–2.99 (m, 6H), 2.91–2.82 (m, 2H), 2.67–2.54 (m, 2H), 2.03–1.96 (m, 1H), (three resonances obscured or overlapping); LCMS (ESI) m/z 979 [M + H]+.
N-(4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)benzyl)-4-((2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)butanamide (36)
The title compound was prepared from amine 10 and acid 35a according to general procedure as a yellow solid (10.2 mg, 41%); 1H NMR (400 MHz, DMSO-d6) δ 11.09 (s, 1H), 10.73 (s, 1H), 9.68 (s, 1H), 8.38 (dd, J = 8.0 and 5.2 Hz, 1H), 8.11 (d, J = 9.2 Hz, 1H), 7.57–7.54 (m, 3H), 7.40 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.20–7.16 (m, 4H), 7.09 (d, J = 8.8 Hz, 1H), 7.03–7.01 (m, 3H), 6.94 (t, J = 7.2 Hz, 1H), 6.83 (d, J = 8.8 Hz, 2H), 6.65–6.62 (m, 1H), 5.04 (dd, J = 5.2 and 12.8 Hz, 1H), 4.22 (d, J = 5.6 Hz, 2H), 4.09 (dd, J = 8.4 and 16.4 Hz, 1H), 3.72–3.70 (m, 4H), 3.06–3.02 (m, 4H), 2.93–2.80 (m, 2H), 2.67–2.50 (m, 3H), 2.25 (t, J = 7.2 Hz, 2H), 2.07–2.00 (m, 1H), 1.88–1.80 (m, 2H), 1.26–1.23 (m, 2H); LCMS (ESI) m/z 875 [M + H]+.
N-(4-(N-((S)-3-(1H-Indol-3-yl)-1-((4-morpholinophenyl)amino)-1-oxopropan-2-yl)sulfamoyl)benzyl)-10-((2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)decanamide (37)
The title compound was prepared from amine 10 and acid 35b according to general procedure as a yellow solid (7.8 mg, 26%); 1H NMR (400 MHz, DMSO-d6) δ 11.08 (s, 1H), 10.72 (s, 1H), 9.68 (s, 1H), 8.27 (dd, J = 6.4 and 5.4 Hz, 1H), 8.11 (brs, 1H), 7.59–7.56 (m, 3H), 7.39 (d, J = 8.4 Hz, 1H), 7.28 (d, J = 7.6 Hz, 1H), 7.17 (d, J = 8.0 Hz, 4H), 7.08 (d, J = 8.8 Hz, 1H), 7.05–7.00 (m, 3H), 6.93 (t J = 7.2 Hz, 1H), 6.83 (d, J = 8.8 Hz, 2H), 6.53–6.50 (m, 1H), 5.04 (dd, J = 5.2 and 12.4 Hz, 1H), 4.21 (d, J = 6.0 Hz, 2H), 4.09 (brs, 1H), 3.73–3.70 (m, 4H), 3.07–3.01 (m, 4H), 2.92–2.80 (m, 2H), 2.67–2.56 (m, 3H), 2.12 (t, J = 7.2 Hz, 2H), 2.02–1.94 (m, 2H), 1.56–1.51 (m, 4H), 1.29–1.25 (m, 11H); LCMS (ESI) m/z 959 [M + H]+.
(2S)-2-((4-((3–(2-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)propanamido)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (39)
The title compound was prepared from amine 10 and acid 38a according to general procedure as a yellow solid (5.8 mg, 23%); 1H NMR (400 MHz, DMSO-d6) δ 11.12 (s, 1H), 10.75 (s, 1H), 9.71 (s, 1H), 8.39 (dd, J = 6.4 and 5.2 Hz, 1H), 8.15 (brs, 1H), 7.59–7.55 (m, 3H), 7.40 (d, J = 8.4 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.19–7.11 (m, 5H), 7.05–7.01 (m, 3H), 6.93 (dd, J = 7.2 and 7.6 Hz, 1H), 6.83 (d, J = 8.4 Hz, 2H), 6.60–6.59 (m, 1H), 5.05 (dd, J = 5.6 and 12.4 Hz, 1H), 4.22 (d, J = 5.6 Hz, 2H), 4.10–4.06 (m, 1H), 3.71–3.66 (m, 6H), 3.58–3.56 (m, 3H), 3.45–3.44 (m, 2H), 3.06–3.01 (m, 4H), 2.89–2.81 (m, 2H), 2.67–2.59 (m, 2H), 2.43–2.40 (m, 2H), 2.04–1.97 (m, 1H); LCMS (ESI) m/z 905 [M + H]+.
(2S)-2-((4-((3–(2–(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)propanamido)methyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (40)
The title compound was prepared from amine 10 and acid 38b according to general procedure as a yellow solid (3.9 mg, 15%); 1H NMR (400 MHz, DMSO-d6) δ 11.08 (s, 1H), 10.72 (s, 1H), 9.68 (s, 1H), 8.35–8.33 (dd, J = 6.4 and 5.2 Hz, 1H), 8.11 (brs, 1H), 7.59–7.56 (m, 3H), 7.40 (d, J = 8.4 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.20–7.11 (m, 5H), 7.04–7.01 (m, 3H), 6.93 (dd, J = 8.0 and 6.4 Hz, 1H), 6.82 (d, J = 8.8 Hz, 2H), 6.60–6.58 (m, 1H), 5.05 (dd, J = 5.6 and 13.2 Hz, 1H), 4.23 (d, J = 6.0 Hz, 2H), 4.09 (brs, 1H), 3.72–3.70 (m, 4H), 3.66–3.59 (m, 4H), 3.55–3.46 (m, 4H), 3.45–3.44 (m, 3H), 3.03–3.00 (m, 4H), 2.86–2.80 (m, 2H), 2.67–2.60 (m, 2H), 2.40–2.37 (m, 2H), 2.05–1.97 (m, 1H); LCMS (ESI) m/z 949 [M + H]+.
(2S)-2-((4–(14-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3-oxo-6,9,12-trioxa-2-azatetradecyl)phenyl)sulfonamido)-3-(1H-indol-3-yl)-N-(4-morpholinophenyl)propanamide (41)
The title compound was prepared from amine 10 and acid 38c according to general procedure as a yellow solid (13.7 mg, 49%); 1H NMR (400 MHz, DMSO-d6) δ 11.08 (s, 1H), 10.72 (s, 1H), 9.67 (s, 1H), 8.45 (dd, J = 6.4 and 5.2 Hz, 1H), 8.10 (brs, 1H), 7.59–7.56 (m, 3H), 7.40 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 7.6 Hz, 1H), 7.20–7.12 (m, 5H), 7.04–7.01 (m, 3H), 6.93 (dd, J = 8.0 and 6.8 Hz, 1H), 6.83 (d, J = 8.8 Hz, 2H), 6.60–6.58 (m, 1H), 5.05 (dd, J = 5.2 and 13.2 Hz, 1H), 4.23 (d, J = 5.6 Hz, 2H), 4.10 (brs, 1H), 3.73–3.70 (m, 4H), 3.64–3.59 (m, 4H), 3.54–3.43 (m, 11H), 3.07–3.01 (m, 4H), 2.92–2.80 (m, 2H), 2.60–2.50 (m, 2H), 2.40–2.36 (m, 2H), 2.03–1.97 (m, 1H); LCMS (ESI) m/z 993 [M + H]+.
(2S)-N-(4–(4-(4-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)butanoyl)piperazin-1-yl)phenyl)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamide (44)
The title compound was prepared from tosyl-L-tryptophan (42) and 43a according to general procedure as an orange solid (7 mg, 43%). 1H NMR (400 MHz, DMSO-d6) δ 11.09 (brs, 1H), 10.76 (s, 1H), 9.67 (s, 1H), 8.06 (brs, 1H), 7.58 (t, J = 8.0 Hz, 1H), 7.50 (d, J = 8.0 Hz, 2H), 7.39 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.21–7.17 (m, 3H), 7.10–7.07 (m, 3H), 7.05–7.01 (m, 2H), 6.92 (t, J = 7.2 Hz, 1H), 6.84 (d, J = 8.4 Hz, 2H), 6.68 (brs, 1H), 5.05 (dd, J = 5.2 and 13.2 Hz, 1H), 4.07 (brs, 1H), 3.56 (brs, 4H), 3.04–3.01 (m, 5H), 2.92–2.80 (m, 3H), 2.67–2.56 (m, 2H), 2.47–2.43 (m, 2H), 2.23 (s, 3H), 2.01 (brs, 2H), 1.83–1.82 (m, 2H); LCMS (ESI) m/z 859 [M + H]+.
(2S)-N-(4–(4-(3–(2-((2–(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)propanoyl)piperazin-1-yl)phenyl)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamide (45)
The title compound was prepared from tosyl-L-tryptophan (42) and 43b according to general procedure as a yellow solid (13 mg, 31%). 1H NMR (400 MHz, DMSO-d6) δ 11.10 (brs, 1H), 10.75 (s, 1H), 9.66 (s, 1H), 8.08 (brs, 1H), 7.57 (t, J = 8.0 Hz, 1H), 7.50 (d, J = 8.4 Hz, 2H), 7.40 (d, J = 8.8 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.8 Hz, 2H), 7.13 (d, J = 8.8 Hz, 1H), 7.10–6.97 (m, 5H), 6.92 (t, J = 7.2 Hz, 1H), 6.80 (d, J = 8.8 Hz, 2H), 6.58–6.57 (m, 1H), 5.05 (dd, J = 6.7 and 13.9 Hz, 1H), 4.07 (brs, 1H), 3.70 (t, J = 6.4 Hz, 2H), 3.61–3.57 (m, 4H), 3.47–3.32 (m, 2H), 3.06–2.98 (m, 5H), 2.92–2.80 (m, 2H), 2.64–2.62 (m, 4H), 2.61–2.53 (m, 2H), 2.19 (s, 3H), 2.2–1.99 (m, 1H); LCMS (ESI) m/z 889 [M + H]+.
(2S)-N-(4–(4-(3–(2–(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)propanoyl)piperazin-1-yl)phenyl)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamide (46)
The title compound was prepared from tosyl-L-tryptophan (42) and 43c according to general procedure as a yellow solid (19 mg, 51%). 1H NMR (400 MHz, DMSO-d6) δ 11.10 (brs, 1H), 10.76 (s, 1H), 9.66 (s, 1H), 8.08 (brs, 1H), 7.57 (t, J = 8.0 Hz, 1H), 7.50 (d, J = 8.4 Hz, 2H), 7.40 (d, J = 8.8 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.19 (d, J = 8.8 Hz, 2H), 7.13 (d, J = 8.8 Hz, 1H), 7.10–6.97 (m, 5H), 6.92 (t, J = 7.6 Hz, 1H), 6.82 (d, J = 8.8 Hz, 2H), 6.62–6.59 (m, 1H), 5.05 (dd, J = 6.7 and 13.9 Hz, 1H), 4.07 (brs, 1H), 3.65–3.53 (m, 10H),3.45–3.44 (m, 4H), 3.06–2.99 (m, 5H), 2.91–2.84 (m, 2H), 2.59–2.53 (m, 4H), 2.23 (s, 3H), 2.2–1.99 (m, 1H); LCMS (ESI) m/z 933 [M + H]+.
(2S)-N-(4–(4-(3–(2–(2–(2-((2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)ethoxy)ethoxy)ethoxy)propanoyl)piperazin-1-yl)phenyl)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamide (47)
The title compound was prepared from tosyl-L-tryptophan (42) and 43d according to general procedure as an orange solid (5 mg, 37%). 1H NMR (400 MHz, DMSO-d6) δ 11.07 (brs, 1H), 10.75 (s, 1H), 9.68 (s, 1H), 8.05 (brs, 1H), 7.57 (t, J = 8.0 Hz, 1H), 7.50 (d, J = 8.0 Hz, 2H), 7.39 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 7.6 Hz, 1H), 7.19 (d, J = 9.2 Hz, 2H), 7.13 (d, J = 8.0 Hz, 1H), 7.14–7.01 (m, 5H), 6.91 (t, J = 8.0 Hz, 1H), 6.83 (d, J = 8.0 Hz, 2H), 6.60 (m, 1H), 5.05 (dd, J = 5.2 and 12.8 Hz, 1H), 4.06 (brs, 1H), 3.64–3.45 (m, 14H), 3.04 (m, 3H), 2.99 (m, 2H), 2.89–2.80 (m, 3H), 2.60–2.57 (m, 6H), 2.23 (s, 3H), 2.03 (brs, 2H); LCMS (ESI) m/z 977 [M + H]+.
(2S)-N-(4–(4-(1-((2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3,6,9,12-tetraoxapentadecan-15-oyl)piperazin-1-yl)phenyl)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamide (48)
The title compound was prepared from tosyl-L-tryptophan (42) and 43e according to general procedure as a pale-yellow solid (5.0 mg, 25%). 1H NMR (400 MHz, Acetone-d6) δ 9.97 (brs, 1H), 9.90 (brs, 1H), 8.90 (brs, 1H), 7.60–7.54 (m, 3H), 7.41 (d, J = 7.6 Hz, 1H), 7.33–7.30 (m, 3H), 7.14–7.09 (m, 4H), 7.07–7.04 (m, 2H), 6.94 (t, J = 8.0 Hz, 1H), 6.87 (d, J = 8.8 Hz, 2H), 6.62 − 6.59 (m, 2H), 5.07 (dd, J = 5.4 and 12.6 Hz, 1H), 4.16–4.11 (m, 1H), 3.86–3.71 (m, 4H), 3.69–3.65 (m, 4H), 3.64–3.61 (m, 4H), 3.58–3.57 (m, 8H), 3.55–3.53 (m, 2H), 3.26–3.21 (m, 1H), 3.14–3.09 (m, 2H), 3.07–3.03 (m, 2H), 3.00–2.92 (m, 1H), 2.76–2.72 (m, 1H), 2.64 (t, J = 6.4 Hz, 2H), 2.26 (s, 3H), 2.24–2.19 (m, 1H), 2.17–2.09 (m, 1H), 1.61–1.59 (m, 1H); LCMS (ESI) m/z 1019 [M-H]-.
(2S)-N-(4–(4-(3-((2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)-3-oxopropyl)piperazin-1-yl)phenyl)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamide (MC_009279) (51)
The title compound was prepared from tosyl-L-tryptophan (42) and 3–(4-(4-aminophenyl)piperazin-1-yl)-N-(2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)propanamide (50a) according to general procedure as a yellow solid (37.5 mg, 55%). 1H NMR (400 MHz, DMSO-d6) δ 11.12 (brs,1H), 10.75 (s, 1H), 10.38 (s, 1H), 9.63 (s, 1H), 8.55 (d, J = 8.4 Hz, 1H), 8.04 (brs, 1H), 7.84 (dd, J = 7.6 and 8.0 Hz, 1H), 7.61 (d, J = 7.2 Hz, 1H), 7.51 (d, J = 8.4 Hz, 2H), 7.40 (d, J = 7.6 Hz, 1H), 7.29 (d, J = 8.4 Hz, 1H), 7.18 (d, J = 8.8 Hz, 2H), 7.11–7.02 (m, 4H), 6.93 (t, J = 7.6, 1H), 6.80 (d, J = 9.2 Hz, 2H), 5.12 (dd, J = 4.8 and 12.4 Hz, 1H), 4.09–4.07 (m, 1H), 3.11–3.02 (m, 5H), 2.87–2.81 (m, 2H), 2.71–2.58 (m, 8H), 2.24 (s, 3H), 2.04–1.94 (m, 2H), 1.74 (brs, 1H); LCMS (ESI) m/z 845 [M + H]+.
4–(4-(4-((S)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamido)phenyl)piperazin-1-yl)-N-(2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)butanamide (52)
The title compound was prepared from tosyl-L-tryptophan (42) and amine 50b according to general procedure as a yellow solid (27.0 mg, 45%). 1H NMR (400 MHz, DMSO-d6) δ 11.13 (s, 1H), 10.74 (s, 1H), 9.70 (s, 1H), 9.60 (s, 1H), 8.49 (d, J = 8.4 Hz, 1H), 7.98 (d, J = 9.2 Hz, 1H), 7.82 (t, J = 8.0 Hz, 1H), 7.59 (d, J = 7.2 Hz, 1H), 7.51 (d, J = 8.0 Hz, 2H), 7.40 (d, J = 7.6 Hz, 1H), 7.29 (d, J = 8.0 Hz, 1H), 7.16 (d, J = 9.2 Hz, 2H), 7.11–7.02 (m, 4H), 6.92 (t, J = 7.2 Hz, 1H), 6.78 (d, J = 9.2 Hz, 2H), 5.13 (dd, J = 5.2 and 12.4 Hz, 1H), 4.13–4.04 (m, 1H), 3.07–3.01 (m, 5H), 2.94–2.81 (m, 3H), 2.67–2.58 (m, 2H), 2.40 (brs, 2H), 2.23 (s, 3H), 2.09–2.04 (m, 1H), 1.87–1.82 (m, 2H), (five resonance obscured or overlapping); LCMS (ESI) m/z 857 [M-H]-.
5–(4-(4-((S)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamido)phenyl)piperazin-1-yl)-N-(2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)pentanamide (53)
The title compound was prepared from tosyl-L-tryptophan (42) and amine 50c according to general procedure as a yellow solid (7.8 mg, 25%). 1H NMR (400 MHz, DMSO-d6) δ 11.14 (s, 1H), 10.75 (s, 1H), 9.70 (s, 1H), 9.62 (s, 1H), 8.47 (t, J = 8.4 Hz, 1H), 8.04 (brs, 1H), 7.83 (t, J = 8.0 Hz, 1H), 7.62 (d, J = 7.2 Hz, 1H), 7.50 (d, J = 8.0 Hz, 2H), 7.39 (d, J = 7.6 Hz, 1H), 7.28 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 8.8 Hz, 2H), 7.11–7.01 (m, 4H), 6.92 (t, J = 7.2 Hz, 1H), 6.80 (d, J = 9.2 Hz, 2H), 5.14 (dd, J = 5.2 and 12.8 Hz, 1H), 4.09–4.06 (m, 1H), 3.04–3.01 (m, 5H), 2.94–2.86 (m, 2H), 2.67–2.54 (m, 2H), 2.34 (dd, J = 7.2 and 6.8 Hz, 2H), 2.05 (s, 3H), 2.07–2.03 (m, 1H), 1.97–1.92 (m, 1H), 1.70–1.63 (m, 2H), 1.56–1.49 (m, 2H), (five resonances obscured or overlapping); LCMS (ESI) m/z 871 [M-H]-.
6–(4-(4-((S)-3-(1H-indol-3-yl)-2-((4-methylphenyl)sulfonamido)propanamido)phenyl)piperazin-1-yl)-N-(2–(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)hexanamide (54)
The title compound was prepared from tosyl-L-tryptophan (42) and amine 50d according to general procedure as a yellow solid (5.4 mg, 12%). 1H NMR (400 MHz, DMSO-d6) δ 11.14 (s, 1H), 10.75 (s, 1H), 9.70 (s, 1H), 9.61 (s, 1H), 8.48 (t, J = 8.0 Hz, 1H), 8.03 (d, J = 8.0 Hz, 1H), 7.83 (t, J = 8.0 Hz, 1H), 7.61 (d, J = 7.2 Hz, 1H), 7.51 (d, J = 8.0 Hz, 2H), 7.40 (d, J = 8.0 Hz, 1H), 7.28 (d, J = 8.4 Hz, 1H), 7.17 (d, J = 9.2 Hz, 2H), 7.11–7.01 (m, 4H), 6.92 (t, J = 7.2 Hz, 1H), 6.79 (d, J = 9.2 Hz, 2H), 5.15 (dd, J = 5.6 and 12.8 Hz, 1H), 4.10–4.06 (m, 1H), 3.06–3.01 (m, 5H), 2.94–2.80 (m, 2H), 2.67–2.54 (m, 2H), 2.31 (t, J = 7.2 Hz, 2H), 2.23 (s, 3H), 2.08–2.05 (m, 1H), 1.97–1.93 (m, 1H), 1.70–1.62 (m, 2H), 1.53–1.46 (m, 2H), 1.40–1.33 (m, 2H), (five resonances obscured or overlapping); LCMS (ESI) m/z 887 [M + H]+.
Results and discussion
Design and synthesis of AIMP2-DX2 targeting PROTACs
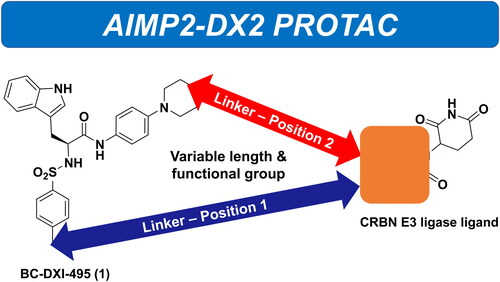
We designed and synthesised AIMP2-DX2 PROTACs using the reported potent small-molecule, BC-DXI-495 (1) as the targeting protein ligase ligand (). Based on the analysis of the docking model of compound 1 in the AIMP2-DX2-GST domain, the toluene part in position 1 and the morpholine part of position 2 have enough space to add linkers since the compound is less sterically hindered and exposed to the surface whereas the indole part of the molecule is embedded inside the proteinCitation9. Accordingly, the addition of various forms of the linkers and E3 ligase ligands were attempted in both positions 1 and 2 to find the best combination of the PROTAC.
Figure 2. Chemical Structures of BC-DXI-495 (1) and putative positions to link the E3 ligase ligand.

Among a slew of E3 ubiquitin ligases, Siah1 protein is known to regulate the level of AIMP2-DX2 in cells using the ubiquitination-degradation processCitation9. Therefore, in order to degrade the target protein AIMP2-DX2 through chemically induced degradation, it is necessary to find an appropriate E3 ligase ligand to bind this Siah1. Currently, two types of E3 ligase ligand are being used and studied extensively, i.e. von-Hippel-Lindau (VHL) (2) and Cereblon (CRBN). In the case of CRBN, 4-hydroxythalidomide (3) and pomalidomide (4-aminothalidomide, {4}) are the most used in research ().
Thus, we used these compounds to find the suitable E3 ligase ligand for Siah1 and conducted the surface plasmon resonance (SPR) assay. Both 4-hydroxythalidomide (3) and pomalidomide (4) showed some degree of binding affinity whereas VHL ligand has not shown any affinity with Siah1 (details are not reported). For further confirmation, biotin-attached 4-hydroxythalidomide (5) and pomalidomide (6) were synthesised (see details in Supplementary Material) and conducted the in vitro pull-down assay to see whether both CRBN types of E3 ligase ligands bind to Siah1. The result from the in vitro binding assay shows that Siah1 protein binds to both types of CRBN E3 ligase ligands but not to AIMP2-DX2 which means that 4-hydroxythalidomide (3) and pomalidomide (4) specifically bind to Siah1. Therefore, we decided to use both CRBN types of E3 ligase ligands for our study ().
Chemistry
The synthetic approaches used to generate AIMP2-DX2 PROTACs using BC-DXI-495 (1) derivatives and two types of CRBN-based E3 ligase ligands are described in Schemes 1–5. Since the length of the linker plays a key role in the potency of PROTAC degraders, we systematically varied the linker length in compounds ranging from 4 to 22 atoms with the objective of determining the optimal linker length. The linker and E3 ligase ligand complexes were either purchased from Sigma-Aldrich® or synthesised based on literaturesCitation16–20. As mentioned above, BC-DXI-495 (1) derivative was synthesised according to the procedure shown in Scheme 1. Starting from commercially available L-tryptophan (7) with 4-cyanobenzenesulfonyl chloride under basic conditions to give compound 8 and subsequent standard amide coupling conditions with 4-morphlinoaniline afforded compound 9. Then, the nitrile moiety was reduced to amine 10 and further reaction with 2-azido-1,3-dimethylimidazolinium hexafluorophosphate in dichloromethane gave the azide product 11. Click reaction of compound 11 with alkyne 12a–dCitation16 and 17a–f afforded 4-hydroxy thalidomide-based PROTACs 13–16 and pomalidomide-based PROTACs 18–23, respectively.
Scheme 1. Synthesis of AIMP2-DX2 PROTACs 13–16, 18–23. Reagents and conditions: (i) 4-cyanobenzenesulfonyl chloride, TEA, THF:H2O, 0–25 °C, overnight; (ii) 4-morphlinoaniline, HATU, DIPEA, DMF, 25 °C, overnight; (iii) LiAlH4, THF, 0–25 °C, overnight; (iv) ADMP, DCM, DMAP, 25 °C overnight; (v) (+)-Sodium L-ascorbate, copper(II) sulphate pentahydrate, DMSO, 25 °C, overnight.

The preparation of AIMP2-DX2 degraders containing linear linkers instead of triazole- containing linkers was synthesised (Scheme 2). Thus, hydrolysis of nitrile 9 under basic conditions gave the corresponding acid 24, which was then further treated with various forms of linker and E3 ligase ligand combinations for standard amination conditions to give PROTACs 25–26, 28–31, and 33–34 (Scheme 2).
Scheme 2. Synthesis of AIMP2-DX2 PROTACs 25, 26, 28–31, and 33–34. Reagents and conditions: (i) KOH, 2-propanol, reflux, 18 h; (ii) HATU, DIPEA, DMF, 25 °C, overnight; (iii) HATU, DIPEA, DMF, 25 °C, overnight; (iv) HATU, DIPEA, DMF, 25 °C, overnight.

For other variations, amine 10 was treated with HATU, DIPEA and pomalidomide-based linkers 35a,b and 38a–c in DMF for overnight at room temperature affording the desired AIMP2-DX2 PROTACs 36–37 and 39–41, respectively.
To prepare position 2 linked PROTACs 44–48, the pomalidomide-based analogues used in Scheme 3 were coupled with 4-(piperazin-1-yl) aniline, followed by another amination reaction with HATU and DIPEA to yield the desired compounds 44–48 (Scheme 4).
Scheme 3. Synthesis of AIMP2-DX2 PROTACs 36, 37, and 39–41. Reagents and conditions: (i), (ii) HATU, DIPEA, DMF, 25 °C, overnight.

Scheme 4. Synthesis of AIMP2-DX2 PROTACs 45–49. Reagents and conditions: (i) p-Toluenesulfonyl chloride, NEt3, THF:H2O (10:1 v/v mixture) 0–25 °C, overnight; (ii) HATU, DIPEA, DCM, 25 °C, 1 h; (iii) 4-(piperazin-1-yl) aniline, HATU, DIPEA, DMF, 60 °C, 4 h.

Scheme 5. Synthesis of AIMP2-DX2 PROTACs 51–54. Reagents and conditions: (i) 4-(piperazin-1-yl) aniline, K2CO3, MECN, reflux, overnight; (ii) compound 42, HATU, DIPEA, DCM, 25 °C, 1 h, overnight.

Lastly, E3 ligase linkers 49a–dCitation20 were alkylated with 4-(piperazin-1-yl) aniline to afford corresponding compounds 50a–d. These compounds were then further treated with compound 42 under basic conditions and gave the desired products 51–54.
We then assessed inhibitory effects of all the designed PROTACs 13–16, 18–23, 25, 26, 28–31, 33, 34, 36, 37, 39–41, 44–48, and 51–54 on the AIMP2-DX2-expressing A549 cell in treatment conditions at 4 µM for 24 h and the results are shown in and Citation2. For comparison, BC-DXI-495 (1) was used as the reference compound to compare the inhibitory activity. To be noted, linker length dependence was observed in AIMP2-DX2 degraders. PROTACs with longer linkers, such as 16, 21, 22, 23, and 31 possessed very poor inhibitory activities, which implied that these longer molecules were less preferred (). In addition, even if the same linker and E3 ligase ligand were used, the activity may vary depending on the location where the target molecule is linked. First, triazole containing 4-hydroxythalidomide-based PROTACs 13–16 were assessed for inhibitory activity. Compound 14 with triazole and 9 atoms in the linker was the most potent in this series with about 10% more potent than BC-DXI-495 (1). In the same series, either reducing or increasing the length of the linker decreases efficacy. These results indicate the optimal linker length is around 13 atoms. Switching to pomalidomide-based triazole-containing products, PROTACs 18–23 were tested against AIMP2-DX2. Interestingly, none of the above compounds have superior activity against AIMP2-DX2 compared to compound 1. We then replaced the triazole linkage with an amine linkage. The 4-hydroxythalidomide-based amine-linked PROTACs 25 and 26 resulted in increased potency with 46.16% and 46.39% inhibition, respectively. However, when the E3 ligase ligand was changed to pomalidomide compounds 28–34, the potency was dramatically decreased to the lowest of 14.84%. Considering that compounds 26 and 30 have the same length of the carbon atom linkers, it can be concluded that 4-hydroxythalidomide is preferable with the amide linear linkage. Reverse amide products 36–41 were also tested. Compared to compounds 36 and 37, the inhibitory activity of compounds 39–41 was higher. Therefore, we suspect that the presence of oxygen atoms in the linkage is crucial for AIMP2-DX2 degraders.
Table 1. Structures and the % inhibition AIMP2-DX2 values for PROTACs. 
To compare the position effect of the target, combinations of linkers and E3 ligase ligands were linked to position 2 of the target compound. Therefore, % inhibition of AIMP2-DX2 was measured for compounds 44–48, 51–53, and 54 (). Interestingly, AIMP2-DX2 degrader 44 showed more than 50% inhibitory activity although the length of the linker was only four atoms apart. Introducing one ether unit in the linker, the activity of compound 45 was further increased to 51.99%. However, longer linker lengths of the same series also led to a decrease in inhibitory activity similar to those attached to position 1. When carbonyl is presented next to an E3 ligase ligand rather than the target, the inhibitory activity of compounds 51–54 only presents a decrease in inhibitory activity with an increase in linker length. As a result, it was found that position 2 favours a shorter linker length, such as 4–6 atoms in length between the target and the E3 ligase ligand. In addition, like Position 1, the linker at position 2 also shows better activity with oxygen-containing linkers.
Table 2. Structures and the % inhibition AIMP2-DX2 values for PROTACs with different linkers.
To confirm the inhibitory activity of compounds, 14 compounds with > 37% inhibitory activity were submitted for further assay to determine the 50% inhibitory concentration (IC50). Among these PROTACs, compound 45 was the most potent AIMP2-DX2 degrader with an IC50 value of 2.37 µM (). This indicated that the inhibitory activity of the PROTAC compound was slightly improved and comparable to the small-molecule inhibitor itself (1).
Table 3. IC50 value of compounds against AIMP2-DX2.
In vitro bioassays
Since PROTAC compound links the substrate and E3 ligase, AIMP2-DX2, and Siah1 in this study, respectively, we checked whether compound 45 affects the cellular binding of AIMP2-DX2 and Siah1. As a result, it was observed that treatment of compound 45 enhances the endogenous binding of Siah1 to AIMP2-DX2 in a dose-dependent manner (). Then, we also determined the direct effect of compound 45 on binding of two proteins via in vitro pull-down assay (), implying that compound 45 could function as a PROTAC compound for connecting AIMP2-DX2 and Siah1 as a substrate and E3 ligase, respectively.
Figure 5. Compound 45 mediated induction of interaction between Siah1 and AIMP2-DX2. (A) Immunoprecipitation assay showing compound 45-dependent increased binding of two proteins. IP and WCL mean immunoprecipitation and whole cell lysates, respectively. Actin was used as a loading control. (B) In vitro pull-down assay confirming compound 45-mediated direct effect on binding of two proteins. GST proteins were detected by Coomassie staining. EV means empty vector.

In addition, to check whether the PROTAC compound results in the degradation of AIMP2-DX2 via its binding to Siah1, we checked the protein and mRNA level of AIMP2-DX2. Compound 45 specifically decreased the level of AIMP2-DX2 protein, but not mRNA (), suggesting that compound 45 functions in the PROTAC system. The compound also selectively led to the degradation of AIMP2-DX2 without any effect on AIMP2, implying the specificity of compound 45. We further confirmed that the ubiquitination of AIMP2-DX2 was induced by the treatment of compound 45 (). Altogether, it could be concluded that compound 45 acts as a PROTAC compound to degrade oncogenic protein, AIMP2-DX2.
Figure 6. Ubiquitination-mediated degradation of AIMP2-DX2 via compound 45. (A) Compound 45-mediated alteration of AIMP2-DX2 protein and mRNA level. Cells treated with compound 45 were subjected to western blotting (WB) and RT-PCR (RT). (B) Determination of compound 45-dependent ubiquitination of AIMP2-DX2. The cells expressing strep-AIMP2-DX2 were treated with compound 45 and MG132 and subjected to ubiquitination assay. Ub: ubiquitin.

To ensure whether compound 45 ultimately induces the decline of cell viability via degradation of AIMP2-DX2, we determined the anti-proliferative efficacy of compound 45. Compound 45 efficiently reduced cell viability but there was no significant difference in cell viability, when Siah1 was knocked down using its specific si-RNA (), implying that compound 45 utilises Siah1 as the E3 ligase. Compound 45 was further tested in several lung cancer cell lines, Calu6, HCC1588, and H226, showing different protein levels of AIMP2-DX2 and Siah1. Compound 45 efficiently declined the cell viability of only Calu6 which has a high level of both AIMP2-DX2 and Siah1 among the tested cell lines (). Combined together, compound 45 was unveiled to depend on the existence of both substrate and E3 ligase to function as a PROTAC. From the comparison of EC50 with IC50 of compound 45, we also concluded that the anti-proliferative efficacy of the compound was mediated by degradation of AIMP2-DX2.
Figure 7. Dependency of compound 45-mediated anti-proliferative efficacy on level of both AIMP2-DX2 and Siah1. (A) Siah1-dependency of the suppressed cell viability via compound 45. Siah1-knockdowned cells via its specific si-RNA were treated with compound and subjected to MTT assay. The results were shown as a bar graph (left) and the level of proteins were checked by immunoblotting (right). (B) Compound 45-mediated cell viability in lung cancer cell lines with the different level of AIMP2-DX2 and Siah1. Cell viability was determined as above. N.D.: not determined. (A and B) The experiments were independently repeated three times.

Prediction of binding modes of E3 ligases in the Siah1
We have also performed docking of the E3 ligase ligand pomalidomide (4) and its parent compound, thalidomide against the human E3 ubiquitin ligase Siah1 to predict the binding modes of E3 ligase ligands. The selected homo sapiens Siah1 PDB structure was 4CA1Citation21 with RMSD of 1.58 Å and the overall quality based on wwPDB structure validation indicated a good quality x-ray structure. This homodimer protein structure was prepared for docking using Protein Preparation tool where all waters were removed, and the hydrogens were added. Putative binding sites in each chain were predicted using CASTp version 3.0 (Chicago, IL, USA)Citation22. Predicted Pockets 1 and 3 were targeted for docking ().
Figure 8. Binding modes of pomalidomide (green atom-coloured stick) and thalidomide (yellow atom-coloured stick) in Pockets 1 and 3 of the human Siah1 protein (blue ribbon). The electron density surface is shown for Pocket 3.

The program used for docking was Glide, Schrödinger (release version 2021–4), using the XP modeCitation23. The putative binding sites were defined by selected residues constituting Pockets 1 and 3. Selected Pockets 1 and 3 are exposed towards the surface area, which could be simulated that E3 ligase ligands would be connected with the linker and the target binder. The best docking poses were determined based on docking scores as well as binding modes. Based on the docking results, pomalidomide (4) was docked better than thalidomide in each of the designated Pockets 1 and 3. In general, pomalidomide (4) interacted more with residues due to its additional amino group (see Supplementary Material for more details, Figure S1). This is in correlation with the SPR results where pomalidomide (4) was shown to be the most favourable E3 ligase of choice for Siah1. Overall, E3 ligase ligands docked better in Pocket 3.
Biological methods
Cell culture and materials
The H460, Calu6, HCC1588, H226, A549, and 293 T cell lines were kindly gifted from Biocon (Medicinal Bioconvergence Research Centre, Yonsei University, Seoul, South Korea). The H460, Calu6, HCC1588, H226, and A549 cells were cultivated in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin in 5% CO2 at 37 °C. 293 T cells were cultured in Dulbecco’s modified Eagle’s medium under the same conditions as above. AIMP2-DX2 was cloned into the EcoRI/XhoI sites of the pEXPR-IBA5 vector to express strep-tagged AIMP2-DX2. MG-132 (#474790) was purchased from Millipore. Specific antibodies against AIMP2-DX2 and AIMP2 were purchased from Curebio. The anti-Siah1 (#ab2237), -actin (#A1978), and ubiquitin (#sc-8017) antibodies were purchased from Abcam (Cambridge, UK), Sigma (St. Louis, MO), and Santa Cruz Biotechnology (Dallas, TX).
Screening
A549 cells (4 × 104 cells/well) expressing luciferase-tagged AIMP2-DX2 were seeded in 96-well, flat-bottom white plates (#3903 Corning) and incubated for 12 h. The cells were treated with the compounds (4 μM) for 24 h, and luminescence was detected following the manufacturer’s protocol (Promega, Madison, WI). All experiments were independently repeated three times.
Cell viability assay
Calu6, HCC1588, and H226 (4 × 104 cells/well) cells were cultured in 96-well, flat-bottom plates for 24 h. Serum-free media containing the diluted compounds was added to the cells and incubated for 96 h. Of 10 μL of MTT solution (5 mg/mL, Sigma, St. Louis, MO) was added to the cells and incubated for 1.5 h at 37 °C. After discarding the culture media with the MTT solution, the precipitated formazan crystals in each well were dissolved with 100 μL of DMSO (Duchefa, Haarlem, Netherlands). Absorbance was measured at 560 nm using a microplate reader (Sunrise, TECAN, Männedorf, Switzerland). All experiments were independently repeated three times.
Ubiquitination assay
The 293 T cells expressing strep-tagged AIMP2-DX2 were treated with the compounds with MG-132 (50 μM) and lysed with 50 mM Tris–HCl (pH 7.4) lysis buffer containing 100 mM NaCl, 0.5% Triton X-100, 0.5% SDS, 10% glycerol, 1 mM EDTA, and protease inhibitor cocktail (Calbiochem). AIMP2-DX2 from total cell extracts were precipitated by using strep-tactin column. The precipitates were subjected to SDS-PAGE and immunoblotting using specific antibodies against the proteins of interest.
Conclusions
In summary, a focussed library of AIMP2-DX2 degraders was developed. Various factors, including linker length, functional groups on the linker, type of E3 ligase ligands and target connection positions were systematically evaluated to understand AIMP2-DX2 PROTACs for deriving the best strategy for inhibiting AIMP2-DX2. As a result, the PROTAC compound 45 targeting AIMP2-DX2 was identified. To the best of our knowledge, compound 45 is the first reported AIMP2-DX2 targeting PROTAC compound. Our data confirmed the applicability of PROTAC technology to target AIMP2-DX2, and this discovery of AIMP2-DX2 PROTAC compound 45 could potentially lead to future therapeutic opportunities to treat lung cancer.
Supplemental Material
Download PDF (1.9 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat Rev Cancer. 2011;11(10):708–718.
- Kwon NH, Fox PL, Kim S. Aminoacyl- tRNA synthetases as therapeutic targets. Nat Rev Drug Discov. 2019;18(8):629–650. −
- Han JM, Lee MJ, Park SG, Lee SH, Razin E, Choi E-C, Kim S. Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem. 2006;281(50):38663–38667.
- Sivaraman A, Kim DG, Bhattarai D, Kim M, Lee HY, Lim S, Kong J, Goo J-I, Shim S, Lee S, et al. Synthesis and structure–activity relationships of arylsulfonamides as AIMP2- DX2 inhibitors for the development of a novel anticancer therapy. J Med Chem. 2020;63(10):5139–5158.
- Choi JW, Kim DG, Lee AE, Kim HR, Lee JY, Kwon NH, Shin YK, Hwang SK, Chang SH, Cho MH, et al. Cancer-associated splicing variant of tumor suppressor AIMP2/p38: pathological implication in tumorigenesis. PLoS Genet. 2011;7(3):e1001351.
- Jung JY, Kim EY, Kim A, Chang J, Kwon NH, Moon Y, Kang EJ, Sung JS, Shim H, Kim S, et al. Ratio of autoantibodies of tumor suppressor AIMP2 and its oncogenic variant is associated with clinical outcome in lung cancer. J Cancer. 2017;8(8):1347–1354.
- Choi JW, Lee JW, Kim JK, Jeon HK, Choi JJ, Kim DG, Kim BG, Nam DH, Kim HJ, Yun SH, et al. Splicing variant of AIMP2 as an effective target against chemoresistant ovarian cancer. J Mol Cell Biol. 2012;4(3):164–173.
- Kim DG, Choi Y, Lee Y, et al. AIMP2-DX2 provides therapeutic interface to control KRAS-driven tumorigenesis. Nat Commun. 2022;13:2572.
- Lim S, Cho HY, Kim DG, Roh Y, Son SY, Mushtaq AU, Kim M, Bhattarai D, Sivaraman A, Lee Y, et al. Targeting the interaction of AIMP2-DX2 with HSP70 suppresses cancer development. Nat Chem Biol. 2020;16(1):31–41.
- Lee HS, Kim DG, Oh YS, Kwon NH, Lee JY, Kim D, Park SH, Song JH, Lee S, Han JM, et al. Chemical suppression of an oncogenic splicing variant of AIMP2 induced tumour regression. Biochem J. 2013;454(3):411–416.
- Oh AY, Jung YS, Kim J, Lee JH, Cho JH, Chun HY, Park S, Park H, Lim S, Ha NC, et al. Inhibiting DX2-p14/ARF Interaction exerts antitumor effects in lung cancer and delays tumor progression. Cancer Res. 2016;76(16):4791–4804.
- Lee S, Kim DG, Kim K, Kim T, Lim S, Kong H, Kim S, Suh Y-G. 2-Aminophenylpyrimidines as novel inhibitors of aminoacyl-tRNA synthetase interacting multifunctional protein 2 (AIMP2)-DX2 for lung cancer treatment. J Med Chem. 2020;63(8):3908–3914.
- Lee BRa, Gyu Kim D, Mi Kim Y, Kim S, Choi I. Discovery of benzodioxane analogues as lead candidates of AIMP2-DX2 inhibitors. Bioorg Med Chem Lett. 2022;73:128889.
- Lai AC, Crews CM. Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov. 2017;16(2):101–114.
- Wang C, Zhang Y, Wang J, Xing D. VHL-based PROTACs as potential therapeutic agents: recent progress and perspectives. Eur J Med Chem. 2022;227:113906.
- Wurz RP, Dellamaggiore K, Dou H, Javier N, Lo M-C, McCarter JD, Mohl D, Sastri C, Lipford JR, Cee VJ. A “bro” for the rapid synthesis of bispecific molecules for inducing protein degradation. J Med Chem. 2018;61(2):453–461.
- Zhou B, Hu J, Xu F, Chen Z, Bai L, Fernandez-Salas E, Lin M, Liu L, Yang CY, Zhao Y, et al. Discovery of small-molecule degrader of bromodomain and extraterminal (BET) protein with picomolar cellular potencies and capable of achieving tumor regression. J Med Chem. 2018;61(2):462–481.
- Lohbeck J, Miller AK. Practical synthesis of a phthalimide-based cereblon ligand to enable PROTAC development. Bioorg Med Chem Lett. 2016;26(21):5260–5262.
- Qiu X, Sun N, Kong Y, Li Y, Yang X, Jiang B. Chemoselective synthesis of lenalidomide-based PROTAC library using alkylation reaction. Org Lett. 2019;21(10):3838–3841.
- Li W, Gao C, Zhao L, Yuan Z, Chen Y, Jiang Y. Phthalimide conjugations for the degradation of oncogenic PI3K. Eur J Med Chem. 2018;151:237–247.
- Rimsa V, Eadsforth TC, Hunter WN. Two high-resolution structure of the human E3 ubiquitin ligase Siah1. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69(Pt 12):1339–1343.
- Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018; 46(W1):W363–W367.
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complex. J Med Chem. 2006;49(21):6177–6196.