ABSTRACT
Introduction
The microbiome is known to have a substantial impact on human health and disease. However, the impacts of the microbiome on immune system development, susceptibility to infectious diseases, and vaccine-elicited immune responses are emerging areas of interest.
Areas covered
In this review, we provide an overview of development of the microbiome during childhood. We highlight available data suggesting that the microbiome is critical to maturation of the immune system and modifies susceptibility to a variety of infections during childhood and adolescence, including respiratory tract infections, Clostridioides difficile infection, and sexually transmitted infections. We discuss currently available and investigational therapeutics that have the potential to modify the microbiome to prevent or treat infections among children. Finally, we review the accumulating evidence that the gut microbiome influences vaccine-elicited immune responses among children.
Expert opinion
Recent advances in sequencing technologies have led to an explosion of studies associating the human microbiome with the risk and severity of infectious diseases. As our knowledge of the extent to which the microbiome influences childhood infections continues to grow, microbiome-based diagnostics and therapeutics will increasingly be incorporated into clinical practice to improve the prevention, diagnosis, and treatment of infectious diseases among children.
1. Introduction
The human microbiota is comprised of trillions of microbes – including bacteria, fungi, viruses, and archaea – that have co-evolved with our species for millennia. The number of microbes colonizing our bodies is truly staggering; for instance, infants have an estimated 1.9 trillion human cells but are colonized by more than twice that number of microbial cells, including approximately 4.4 trillion bacterial cells [Citation1]. The human microbiome, which refers to the collection of microbial genomes harbored by this microbiota, is equally vast; in an analysis of 3,655 oral and fecal samples across 13 studies, investigators identified more than 45 million unique bacterial genes harbored by these individuals, approximately half of which were unique to a single sample [Citation2]. By mass, most of the microbes that colonize our bodies reside within the gastrointestinal tract, but complex microbial communities also inhabit the entirety of the aerodigestive tract, skin, and genitourinary tract. Increasingly, this human microbiota is recognized to play critical roles in the maintenance of health and protection from infections, particularly during childhood. In this review, we provide an overview of the factors that influence microbiome development during childhood, summarize the available data regarding the impact of the microbiota on the risk and severity of childhood infections, and discuss the potential utility of microbiome-based therapies to prevent or treat infections among children.
1.1. Development of the microbiome during childhood
At no point across the lifespan is the microbiome as dynamic as during childhood. At birth, infants are colonized by trillions of pioneering microbes from the maternal vaginal, gut, and skin microbiota. Over the ensuing days and months, these microbial communities undergo substantial shifts in composition due to changes in the local microenvironment, environmental exposures, and microbial competition. For instance, shortly after birth, the gut is an aerobic environment typically containing high abundances of facultative anaerobes, including bacteria from the genera Enterobacter, Enterococcus, and Escherichia [Citation3]. In the subsequent days, the gut becomes increasingly devoid of oxygen, leading to an increase in the abundances of Clostridium and other strictly anaerobic bacteria [Citation4]. The gut microbiota then enters a transitional phase characterized by high abundances of Bifidobacterium species, particularly among breastfed infants, before becoming enriched with Bacteroides, Clostridium, and Ruminococcus following the introduction of solid foods [Citation3–5]. Although typical developmental patterns have been described for microbial communities at several anatomical sites during early life, there is an enormous amount of variability in the composition of these microbiomes as they mature across individuals.
1.2. Factors that influence microbiome composition during childhood
Although recent studies suggest that host genetics influences the microbiota during childhood [Citation6,Citation7], the overwhelming majority of the observed variation in microbiota composition results from environmental exposures (). During infancy, delivery mode, feeding practices, and antibiotic exposures have the largest impact on microbiota development. In a longitudinal study of the gut microbiomes of nearly 600 infants in the United Kingdom, Shao and colleagues demonstrated that infants born via Caesarian section (C-section) had disrupted acquisition of maternally derived strains of Bacteroides, a bacterial genus that includes potentially beneficial (commensal) species, and more frequent high-abundance colonization by Enterococcus and other potential pathogens than vaginally delivered infants [Citation8]. Similarly, in a study of 102 Dutch infants, Bosch and colleagues demonstrated that C-section delivery was associated with a delay in upper respiratory tract colonization by the health-associated bacteria Corynebacterium and Dolosigranulum [Citation9]. Delivery mode has also been shown to influence microbiota composition during infancy at other anatomical sites, including skin and the oral cavity [Citation10,Citation11]. Additionally, numerous studies have described the impact of breastfeeding on gut microbiota composition during infancy. In a study of more than 1,000 one-month-old infants, Barnett and colleagues found that an increasing proportion of feeds containing breast milk was associated with higher relative abundances of Bifidobacterium and Lactobacillus, both of which contain keystone species that broadly influence the gut microbial community [Citation12]. Interestingly, Holst and colleagues found that supplementation of infant formula with five different human milk oligosaccharides was associated with a shift in the gut microbiota composition of formula-fed infants such that it more closely resembled that of breastfed infants [Citation13]. Thus, breastmilk appears to have dose-dependent beneficial effects on the gut microbiota that can be replicated to some extent in formula-fed infants through the administration of specific prebiotics. Finally, early-life antibiotic exposures are recognized to have profound and lasting effects on the microbiota. In a recent trial of three commonly prescribed antibiotic regimens among 147 Dutch neonates, antibiotic exposures were associated with decreased gut abundances of several potentially beneficial genera, including Bacteroides and Bifidobacterium, and increased gut abundances of genera containing common pathogens, including Enterococcus and Klebsiella [Citation14]. Interestingly, a variable degree of gut microbiota disruption was observed across the different antibiotic regimens, and alterations of the gut microbiota were found to persist for at least 1 year after antibiotic exposure [Citation14]. Antibiotics also influence the composition of microbial communities at other anatomical sites; in a longitudinal study of the nasopharyngeal microbiomes of 179 healthy infants in Botswana, Kelly and colleagues found that antibiotic exposures were associated with losses of several generally beneficial genera (Corynebacterium, Lactobacillus) and gains of genera containing common respiratory pathogens (Haemophilus, Moraxella) [Citation15].
Figure 1. Host and environmental factors that influence human microbiome composition. Diet has a major influence on the composition of human microbial communities across the lifespan. Delivery mode is among the most important influences on the microbiota during infancy. Other environmental exposures, such as antibiotic treatment or daycare attendance, can have profound, short-term effects on microbiota composition. Host genetics influences microbiota composition at all ages, albeit to a far lesser extent than environmental exposures.
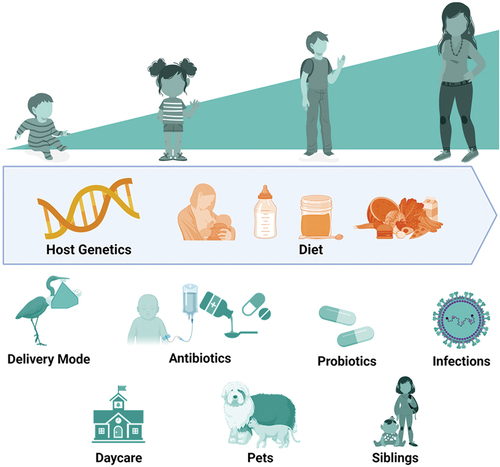
The comparatively few pediatric microbiome studies that have been conducted after infancy suggest that the microbiota continues to develop and mature in response to environmental exposures. In particular, increasing age has consistently been associated with changes in microbiota composition during toddlerhood. During the second and third years of life, the gut microbiota increases in diversity (number of species), and a gradual loss of Bifidobacterium species is typically observed; together, these shifts over time result in the gut microbiota of older children more closely resembling that of adults [Citation16,Citation17]. Diet and nutrition also continue to have substantial effects on the microbiota after infancy and likely contribute to changes in microbiota composition observed with age. This has perhaps been demonstrated most profoundly by studies of the microbiomes of children experiencing undernutrition. The gut microbiomes of children with malnutrition or stunting have been shown to be developmentally immature, with composition resembling children of younger chronological age [Citation18,Citation19]. Moreover, this phenomenon can be effectively reversed by complementary foods designed to enrich for specific bacterial species in the gut microbiota. For example, in a double-blind, randomized controlled trial of such a microbiota-directed complementary food, the gut microbiota of 63 children with moderate acute malnutrition demonstrated improved maturity and enrichment by several bacterial species that typically increase in abundance during weaning [Citation20]. Finally, antibiotic exposures can have long-term impacts on microbiota composition after infancy. In a study of preschool and school-aged Finnish children, the gut microbiota of children who received penicillin recovered in 6–12 months, whereas the gut microbiota of children who received macrolide therapy had still not fully recovered 2 years after antibiotic exposure [Citation21]. The effect of antibiotic exposures on the gut microbiota of children likely varies based on the agent and duration of treatment, and future studies are needed to quantify the degree and duration of microbiota disruption associated with specific antibiotic exposures.
Other environmental factors have been associated with alterations to the microbiota during childhood. Among 33 Japanese children, those who attended daycare were less likely to have nasopharyngeal microbiota profiles dominated by the health-associated species Corynebacterium propinquum [Citation22]. Similarly, the gut microbiomes of 61 children attending daycare differed from those of 24 home-schooled children, with daycare attendance being associated with enrichment of the gut microbiota with several bacteria that were more abundant in older children and adults [Citation23]. Finally, several other environmental factors were associated with gut microbiota composition during the first 3 years of life in a study of 903 children from the United States and Western Europe, including maternal body mass index, probiotic exposures, vitamin D supplementation, geographic location, siblings, furry household pets, and living on a farm [Citation24].
1.3. The microbiome and colonization resistance during childhood
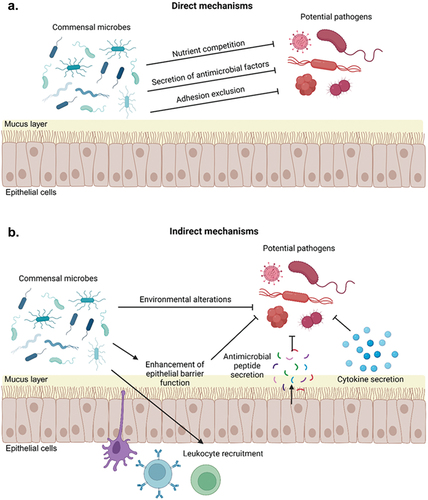
The human microbiota provides protection against infection by preventing colonization and invasion by exogenous pathogens. This important function of the microbiota, referred to as colonization resistance, occurs through a variety of mechanisms, including competition for nutritional sources, production of antimicrobial compounds, alterations of the local microenvironment, and modulation of host immune responses (). In turn, perturbations of the microbiota in response to external factors can lead to a reduction in colonization resistance, rendering children more susceptible to infections. Colonization resistance was first recognized in the 1950s when it was noted that mice that were treated with the antibiotic streptomycin were susceptible to Salmonella infection at oral doses far lower than the typical minimal infectious dose [Citation25]. Subsequent studies have established that the human microbiota similarly resists infections caused by enteric pathogens as well as infections by pathogens at other mucosal surfaces. Moreover, as methods to study microbial interactions have advanced over time, so too has our understanding of the molecular underpinnings of colonization resistance. For instance, using fecal samples collected from children, Osbelt, and colleagues demonstrated that specific strains of Klebsiella oxytoca cultured from these samples effectively prevented or eradicated colonization by a multidrug-resistant strain of Klebsiella pneumoniae in mice [Citation26]. The authors subsequently demonstrated that these protective strains of K. oxytoca more effectively utilized gut carbohydrate sources than the K. pneumoniae strain or nonprotective strains of K. oxytoca, demonstrating that protective K. oxytoca strains resist gut colonization by K. pneumoniae through nutrient competition [Citation26]. Future studies of the molecular mechanisms of colonization resistance have the potential to further our understanding of human microbial ecology and inform the development of microbiome-targeted therapies that prevent or treat childhood infections.
Figure 2. Overview of mechanisms of colonization resistance. Commensal microbes can inhibit colonization and invasion by exogenous pathogens through direct (a) and indirect (b) mechanisms. Direct mechanisms of colonization resistance include competition for space or adhesion sites, nutrient consumption, or secretion of antimicrobial peptides or other factors that directly kill exogenous pathogens. Indirect mechanisms of colonization resistance include altering the local microenvironment so that is it less hospitable to exogenous pathogens and modulation of host immune responses, including by altering cytokine production, recruiting leukocytes to mucosal surfaces, and stimulation production of host antimicrobial peptides by epithelial cells.
1.4. Interactions between microbes and the host immune system during childhood
Early-life colonization of mucosal surfaces by microbes is critical to immune system development [Citation27]. Our understanding of mechanistic relationships between the microbiota and the host immune system is largely derived from experiments conducted using germ-free animals. In these animals, the absence of microbes in early life affects nearly every aspect of the immune system, resulting in deficiencies in mucosal defenses and epithelial cell morphology, altered lymphoid structures, and impaired adaptive immunity [Citation28]. These profound disruptions of immune system development relate to the critical roles that the microbiota plays in immune system education and the development of immune tolerance. Studies that have introduced commensal microbes at various points during the lifespan of germ-free mice have identified windows of opportunity during which exposure to microbes can correct these immune system defects [Citation29]. Notably, early-life microbiota disruption in human infants has been linked to the later development of several immune or inflammatory disorders, including atopic diseases, inflammatory bowel disease, and asthma [Citation30–32]. The predisposition to these conditions may relate to altered cellular immunity, as an absent or altered gut microbiota influences T helper and T regulatory cell functions in mice [Citation33,Citation34]. Taken together, these observations demonstrate that the early-life microbiota primes the immune system for the generation of antigen-specific immune responses to pathogens while simultaneously restraining immune responses to commensal microbes and host antigens.
The human microbiota continues to modulate immune system function throughout the lifespan. In health, microbiome-host homeostasis is characterized by low levels of inflammation and immune cell recruitment sufficient to create a state that is primed to respond to potential pathogens [Citation35,Citation36]. In contrast, alterations of the microbiota can result in a loss of homeostasis and unchecked inflammation and immune activation, contributing to the pathophysiology of diseases such as asthma, ulcerative colitis, and allergic rhinosinusitis [Citation37]. Communication between commensal microbes and the host at mucosal surfaces is primarily mediated through interactions between conserved microbial antigens, referred to as microbial-associated molecular patterns, and pattern recognition receptors (e.g. Toll-like receptors) present on epithelial cells and antigen-presenting cells [Citation38]. Activation of these host receptors initiates innate and adaptive immune responses that are required to maintain homeostasis between the host and the microbiota. Additionally, commensal microbes produce metabolites and other molecules that can directly modulate specific aspects of the host immune system. For instance, some gut bacteria release short-chain fatty acids, secondary bile acids, and other metabolic products that have beneficial effects on intestinal integrity and mucosal immunity [Citation39,Citation40].
The microbiota influences the development of host immune responses to pathogens through several mechanisms. Commensal microbes can induce host epithelial cells to secrete antimicrobial peptides that target specific pathogens, as well as other commensal species [Citation41]. While multiple stimuli can induce epithelial cell secretion of antimicrobial peptides, antibiotic-mediated depletion of the gut microbiota reduces antimicrobial peptide secretion and increases susceptibility to enteric pathogens [Citation42]. Further, microbial interactions with host epithelial and immune cells can modulate cytokine production and inflammation at mucosal surfaces and alter the recruitment and behavior of immune cells. Studies in antibiotic-treated or germ-free mice have identified specific cytokines whose production is induced by the commensal microbiota. For example, Sequeira and colleagues found that Bacteroidetes-elicited interleukin-36 secretion was sufficient to prevent K. pneumoniae colonization of the gut [Citation43]. Similarly, Brown and colleagues demonstrated that microbiota-stimulated secretion of granulocyte-macrophage colony-stimulating factor promoted clearance of respiratory pathogens from the lungs [Citation44]. These studies highlight the multifaceted impact that microbes have on host immune function. While studies in germ-free animals and associative human studies have identified important roles for the microbiota in immune system development, further work is needed to elucidate the molecular mechanisms of these microbe–host interactions and more clearly define such relationships outside of the gut.
2. The microbiome and the risk and severity of childhood infections
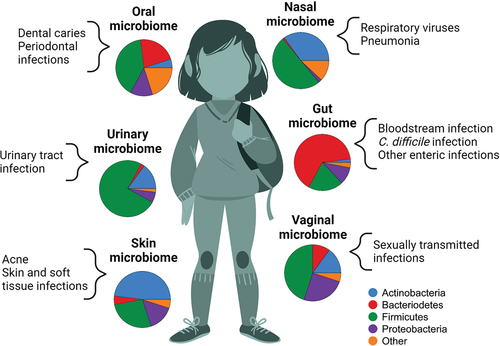
With advances in our knowledge of the factors that disrupt microbiome composition during childhood and the role that the microbiota plays in immune development, there has been substantial interest in identifying microbiome characteristics that are associated with infection susceptibility or severity (). Although most of the research in this area has focused on the upper respiratory and gut microbiomes, microbial communities at other body sites are increasingly recognized to influence the development and course of childhood infections.
Figure 3. The impact of the microbiome on infections during childhood and adolescence. Substantial variability exists in the composition of the microbial communities that colonize children and adolescents at different anatomical sites. The pie charts in this figure depict the relative proportion of sequencing reads assigned to specific bacterial phyla at six body sites. Variability in microbiome composition across individuals at a specific anatomical site contributes to observed differences in infection susceptibility and severity across children.
2.1. The microbiome and childhood respiratory infections
Globally, acute respiratory infections cause more than 740,000 child deaths each year [Citation45]. These infections are preceded by establishment of pathogens within the upper respiratory tract. As described above, the microbiota strongly influences colonization resistance and host immune responses to potential pathogens, and there is growing evidence that the upper respiratory microbiota is a key determinant of respiratory infection susceptibility and severity. Two recent studies evaluated associations between pathogen colonization and microbiota composition to identify potential interspecies interactions that underlie colonization resistance in the upper respiratory tract. Kelly and colleagues evaluated associations between upper respiratory microbiota composition and the risk of colonization by the major human respiratory pathogen Streptococcus pneumoniae (pneumococcus) among infants in Botswana, identifying species of Corynebacterium as playing a major role in pneumococcal colonization resistance [Citation46]. Moreover, they demonstrated that specific strains of Corynebacterium secrete factors that inhibit in vitro growth of S. pneumoniae [Citation46]. Stubbendieck and colleagues similarly identified associations between the presence of Rothia species in the upper respiratory tract and colonization by Moraxella catarrhalis, a leading cause of acute otitis media and bacterial rhinosinusitis [Citation47]. The authors then demonstrated that specific Rothia strains secrete a peptidoglycan endopeptidase that inhibits M. catarrhalis in vitro [Citation47]. Other recent studies demonstrated that nasal administration of commensal bacterial strains can prevent or eradicate upper respiratory colonization by common respiratory pathogens, including Neisseria meningitidis (meningococcus) and Staphylococcus aureus [Citation48,Citation49]. Future mechanistic studies and clinical trials have the potential to unveil additional mechanisms underlying the colonization resistance provided by the upper respiratory microbiota and lead to a new class of probiotics administered nasally for the prevention of respiratory infections.
There are accumulating data that early-life infections alter microbiome composition or function, contributing to differences in colonization resistance and infection susceptibility later in childhood. In a longitudinal study of 114 healthy infants, de Steenhuijsen Piters and colleagues found that asymptomatic viral infections occurring in the first few months of life were associated with a strong upper respiratory mucosal interferon response that coincided with enrichment of the upper respiratory microbiota by genera containing common pathogens (Haemophilus, Moraxella) [Citation50]. Further, infants with early acquisition of these genera had a greater number of respiratory infections in the first year of life than infants that did not become colonized in the first few months of life, suggesting that early encounters with respiratory viruses may alter the host response and create an environment that promotes future infections [Citation50]. Dissanayake and colleagues used a human airway epithelial cell model to demonstrate that rhinovirus infection promotes adhesion of M. catarrhalis to respiratory epithelium, identifying a potential mechanism by which respiratory viruses may influence upper respiratory microbiota composition [Citation51]. Recent studies have similarly identified alterations of the upper respiratory microbiota in the setting of infections caused by other respiratory viruses, including influenza viruses, respiratory syncytial virus (RSV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [Citation52–54]. Finally, early-life colonization by Mycoplasma pneumoniae, a common cause of community-acquired pneumonia, has been associated with a less diverse upper respiratory microbiota, a higher relative abundance of the common respiratory pathogen Haemophilus influenzae, and a history of recurrent respiratory infections among young children [Citation55].
The severity of respiratory infections is also influenced by upper respiratory microbiota composition. A cross-sectional study of children and adolescents found that a microbiota profile dominated by the putatively beneficial bacterial genera Corynebacterium and Dolosigranulum was associated with the absence of respiratory symptoms among individuals with SARS-CoV-2 infection [Citation53]. Similarly, Penela-Sánchez and colleagues reported that children with rhinovirus/enterovirus-associated lower respiratory tract infections requiring intensive care had a less diverse upper respiratory microbiome and higher abundances of Haemophilus than children with asymptomatic infections caused by these viruses [Citation56]. The severity of infections caused by RSV also varies substantially among children, with some data suggesting that microbe–host interactions within the respiratory tract influence RSV disease severity. A cross-sectional study of young children found that microbiota profiles dominated by H. influenzae or Streptococcus were more prevalent both among all children with RSV infection and the subset of these children who required hospitalization compared to healthy controls [Citation54]. Moreover, RSV-infected children with these microbiota profiles had higher peripheral blood expression of genes related to Toll-like receptor signaling and neutrophil and macrophage activation than children with RSV who had other microbiota profiles [Citation54]. Future studies integrating data on the respiratory microbiota and host immune responses are likely to lead to an improved understanding of the extent to which the upper respiratory microbiota affects the severity of respiratory virus infections during childhood.
2.2. The microbiome and childhood infections arising from the gut
Substantial evidence links the gut microbiota to the risk of enteric infections and other infections arising from the gut. Clostridioides difficile infection (CDI) is the prototypical infection arising from a loss of gut colonization resistance. Ling and colleagues reported that CDI in children was associated with higher gut abundances of genera containing common pathogens (e.g. Enterococcus, Escherichia/Shigella, and Klebsiella) and lower abundances of specific gut anaerobes (e.g. Bacteroides, Faecalibacterium, Parabacteroides), suggesting that the former may facilitate C. difficile colonization and the latter may contribute to C. difficile colonization resistance [Citation57]. Notably, enterococci have been observed to be enriched in the gut microbiomes of individuals with CDI in several studies [Citation58,Citation59]. In a recent study that investigated the impact of enterococcal co-colonization on CDI in mice, Smith, and colleagues found that enterococci provide fermentable amino acids that facilitate colonization by C. difficile, while C. difficile toxin-mediated damage to the gut mucosa enhances enterococcal growth, providing mechanistic evidence that the presence of one of these species may promote the fitness of the other species within the human gut [Citation60]. Interestingly, gut colonization by toxigenic C. difficile is common during the first 12 months of life but is typically asymptomatic, while CDI is a relatively common cause of enterocolitis among older children with medical comorbidities or recent antibiotic exposure [Citation61,Citation62]. The mechanisms underlying the low incidence of severe CDI among infants have not been defined, with both the gut microbiota and host immunity postulated to play roles. Infants colonized by toxigenic strains of C. difficile were reported to develop robust anti-toxin humoral immune responses, but the extent to which these adaptive immune responses are also present in older children and adults is unknown [Citation63]. Finally, persistent colonization by C. difficile among infants is associated with lower abundances of several gut anaerobes (e.g. Bifidobacterium, Lacticaseibacillus, and Lactobacillus), suggesting that gut microbiota composition influences eradication of C. difficile even after colonization has established [Citation62,Citation64]. Taken together, these studies demonstrate the importance of the gut microbiota to the pathogenesis of CDI in children and identify mechanisms of microbe–microbe and microbe–host interactions that could inform development of novel strategies to prevent CDI.
Bloodstream infections cause substantial morbidity and mortality among several vulnerable populations of children. Among premature infants, late-onset sepsis is frequently caused by gut-derived bacteria, and these infections are often preceded by loss of potentially beneficial species from the gut microbiota and an increase in the gut abundance of the BSI-causative strain [Citation65,Citation66]. Similar gut microbiota alterations have been observed prior to BSI among children undergoing hematopoietic cell transplantation (HCT), among whom antibiotic exposures appear to be the major driving factor for pathogen expansion within the gut prior to BSI onset [Citation67]. These and other such studies suggest that serial metagenomic sequencing of the gut microbiota might be a useful approach to identifying children in these patient populations who are at high risk of BSI. As an early proof of concept of this approach, Margolis and colleagues recently reported that ratios of the abundances of bacterial species in the gut microbiota prior to HCT were of some utility in predicting the risk of children developing bacterial infections after HCT [Citation68]. Additional studies investigating serial metagenomic sequencing for the identification of children at high risk of infections are anticipated in the coming years as the costs of metagenomic sequencing continue to decline and analysis of these data becoming increasingly high-throughput.
The gut microbiota is also the major reservoir of antibiotic-resistant microbes in human populations. Shotgun metagenomic sequencing, which theoretically enables detection of all antibiotic resistance genes present within a sample (referred to as the ‘resistome’), has substantially advanced our ability to study antibiotic resistance during childhood. Several studies of the gut resistome among children have demonstrated that acquisition of antibiotic-resistant bacteria occurs early in life, even in the absence of antibiotic treatment or other recognized risk factors [Citation69,Citation70]. For instance, in a study of nearly 3,000 neonates from seven low- and middle-income countries in Africa and South Asia, extended-spectrum beta-lactamase and carbapenemase genes were detected in rectal swab samples from more than half of neonates [Citation71]. Similarly, in a study of 42 Vietnamese infants, children, and adults, the gut microbiomes of infants were found to harbor more antibiotic resistance genes than those of older age groups [Citation72]. Interestingly, treatment of children with antibiotics has been shown to increase the gut abundances of bacterial genes conferring resistance to that antibiotic class as well as genes conferring resistance to other antibiotic classes, thus further challenging the selection of empirical antibiotics for children with suspected infections who have had recent antibiotic exposures [Citation73,Citation74]. Although these and other studies have improved our understanding of the resistome during childhood, further work is needed before this knowledge can be translated into strategies that prevent antibiotic-resistant infections in children.
2.3. The cervicovaginal microbiome and sexually transmitted infections
Adolescents account for more than half of new sexually transmitted infections (STIs) in the United States [Citation75]. Most notably, Chlamydia trachomatis and Neisseria gonorrhea are more common in adolescents than in any other age group, while individuals 13 to 24 years of age accounted for 19% of new HIV infections between 2017 and 2021 [Citation76,Citation77]. The mucosa of the reproductive tract is the primary entry point for STIs. Although previous work has demonstrated that the microbiota influences STI susceptibility in both the male and female reproductive tracts, the majority of studies conducted to date have focused on the cervicovaginal microbiota. The composition of the cervicovaginal microbiota varies widely across individuals, and is influenced by hormone levels, contraceptive use, sexual behaviors, nutrition, stress, and tobacco use [Citation78]. Among women of reproductive age, the cervicovaginal microbiota is frequently dominated by Lactobacillus species which produce lactic acid and help to maintain the low pH of the cervicovaginal microenvironment [Citation79]. Cervicovaginal microbiomes with high abundances of Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners, or Lactobacillus jensenii have generally been associated with health, while microbiomes with low abundances of these species, higher microbial diversity, and high abundances of Gardnerella, Prevotella, and other anaerobes have often been observed in disease [Citation80]. In particular, the latter changes in cervicovaginal microbiota composition are often associated with an increase in vaginal pH and the presence of vaginal discharge, the hallmarks of bacterial vaginosis [Citation78]. In turn, bacterial vaginosis is associated with increased risks of acquisition of several STIs, including C. trachomatis, Trichomonas vaginalis, and human papillomavirus [Citation81–83]. Though the mechanisms underlying these associations require further study, bacterial vaginosis is a proinflammatory state that is associated with epithelial barrier disruption and impaired mucosal immunity, each of which has the potential to increase STI susceptibility [Citation84]. Interestingly, although bacterial vaginosis does not appear to increase the risk of N. gonorrhoeae acquisition, there are some data that suggest that it may influence symptoms of this STI [Citation85]. In particular, women with asymptomatic gonorrhea frequently have cervicovaginal microbiomes with high abundances of Lactobacillus species, while women with symptomatic infections often have microbiomes similar to those observed in bacterial vaginosis [Citation85].
The composition of the cervicovaginal microbiota has also been associated with the risk of HIV infection. Women with cervicovaginal microbiomes with low abundances of Lactobacillus or who have recently been diagnosed with bacterial vaginosis are at higher risk of acquiring HIV [Citation86,Citation87]. Moreover, the cervicovaginal microbiota has been shown to influence the effectiveness of pre-exposure prophylaxis for HIV. Though antiretroviral-based HIV prophylaxis has been shown to be highly effective in men, studies in women have yielded variable results with both orally and vaginally administered formulations [Citation88–91]. In a study conducted among South African women, Klatt and colleagues found that the vaginal microbiota strongly influenced the efficacy of a vaginally applied antiretroviral microbicide gel used for the prevention of HIV infection [Citation92]. More specifically, tenofovir vaginal gel reduced HIV acquisition by 61% among women with a Lactobacillus-dominant cervicovaginal microbiome, but by only 18% among women with other cervicovaginal microbiome profiles [Citation92]. After observing that vaginal tenofovir concentrations were negatively correlated with the cervicovaginal abundances of Gardnerella vaginalis and other anaerobes, the authors demonstrated that G. vaginalis metabolizes tenofovir more quickly than host cells are able to convert it to its active form, elegantly demonstrating one such mechanism by which microbes can influence the effectiveness of antibiotics and other therapeutics [Citation92].
2.4. The oral microbiome and childhood caries
The oral microbiome is integral to the pathogenesis of early childhood caries, a condition caused by specific bacteria in the supragingival plaque that create an acidic microenvironment that promotes tooth demineralization and decay. Higher abundances of Streptococcus and Veillonella have been observed in the oral microbiomes of children who develop early caries, with some data suggesting that Streptococcus mutans is the primary causative agent [Citation93]. In a study that used extracted teeth from children with severe caries, Kim and colleagues concluded that S. mutans is at the core of caries biofilms and produces an extracellular scaffold that provides a protective barrier to other microbes living within this mixed-species biofilm [Citation94]. However, other microbial species are likely to contribute to the development of caries. For instance, cohort studies from Scandinavia and Japan revealed that fewer than half of children with caries had S. mutans detected in their oral cavities, while the mean relative abundance of S. mutans among 20 Swedish adolescents with caries was 0.16% [Citation95,Citation96]. Finally, Teng and colleagues reported that the abundances of S. mutans and multiple other bacterial species in the salivary and plaque microbiota informed development of a model that was highly accurate in predicting the risk of future dental caries among preschool-aged children [Citation97].
3. Therapeutic manipulation of the child microbiome
With accumulating evidence that the microbiota influences infection risk and severity, there has been substantial interest in the development of microbiome-targeted therapies for the prevention or treatment of infections. Although most studies have been conducted in adults, several such interventions have been studied in infants and children.
3.1. Probiotics
Because the gut microbiota is recognized to play a critical role in the development of late-onset sepsis and necrotizing enterocolitis, there has long been interest in modifying the gut microbiota of premature infants through the administration of probiotics. A recent meta-analysis that included 106 randomized controlled trials of more than 25,000 infants concluded that multiple-strain probiotics, especially when combined with prebiotics, were associated with lower incidences of severe necrotizing enterocolitis and mortality, whereas single-strain probiotics combined with lactoferrin were most effective in lowering the incidence of sepsis [Citation98]. In a study of 123 premature infants (gestational age <32 weeks), Beck and colleagues found that infants who received probiotics frequently transitioned to a gut microbiome profile dominated by the probiotic strain that persisted long after probiotic discontinuation [Citation99]. Similarly, in a randomized, controlled trial of a multiple-strain probiotic containing Lacticaseibacillus rhamnosus and several strains of Bifidobacterium conducted among premature infants, Bifidobacterium strains were found to stably colonize the gut weeks after probiotic discontinuation, whereas long-term engraftment of the Lacticaseibacillus strain was not observed [Citation100]. These studies, which generally contrast with what has been observed in studies of probiotics in adults, suggest that early life may present a unique opportunity for probiotics to have long-term effects on gut microbiota composition, although these effects are likely to differ across probiotic strains [Citation101,Citation102]. Finally, administration of probiotics to infants may also prevent gut colonization by antibiotic-resistant bacteria, with several studies reporting that infants receiving probiotics harbor fewer antibiotic resistance genes than infants not receiving these products [Citation103,Citation104].
Most studies of probiotics in older children have evaluated these products among children with acute diarrhea or for the prevention of antibiotic-associated diarrhea or CDI. Probiotic trials conducted among children with diarrheal illnesses have yielded mixed results, with some studies suggesting modest benefit. For instance, in a study of a probiotic containing a strain of the yeast Saccharomyces boulardii conducted among 112 children with acute diarrhea, children receiving the probiotic had illnesses that were on average 1-day shorter than placebo recipients [Citation105]. In addition, a probiotic containing strains of Lactobacillus helveticus and L. rhamnosus did not alter the clinical courses of 886 children with acute gastroenteritis; moreover, no differences in gut microbiota diversity or composition were observed among probiotic and placebo recipients and only transient colonization by the probiotic strains was seen among children in the treatment group [Citation106]. Use of probiotics for the prevention of antibiotic-associated diarrhea or CDI among children have been active areas of research over the past several decades. In a recent meta-analysis of 33 trials involving 6,352 participants, Guo and colleagues concluded that probiotics more than halved the risk of antibiotic-associated diarrhea among children 0 to 18 years, with nine children needing to be treated for every case prevented [Citation107]. A recent meta-analysis of data from 31 clinical trials involving 8,672 adults and children found that probiotics reduced the risk of CDI among participants receiving antibiotics by 60%; however, this effect was only observed in trials in which the baseline risk of CDI was above 5% [Citation108]. Although the conclusions of these analyses should be interpreted with some caution given the substantial heterogeneity of the included trials with regard to the probiotic strain and the timing and duration of administration, these data do suggest that probiotics can effectively prevent antibiotic-associated diarrhea in children and may additionally be of benefit for pediatric populations that are at very high risk of CDI. Unfortunately, the relatively high numbers needed to treat to prevent these conditions combined with the costs and challenges of administration has limited routine use of probiotics among children receiving antibiotics.
3.2. Fecal microbiota transplantation
Fecal microbiota transplantation (FMT) has been used in the management of recurrent CDI for nearly two decades, although use of this procedure in children has been limited until relatively recently. Fortunately, there have been several recent studies demonstrating that FMT is a safe and effective treatment for children with recurrent CDI. For instance, Nicholson and colleagues recently summarized outcomes of 372 patients aged 11 months to 23 years with recurrent CDI who underwent FMT at 18 pediatric health centers between 2004 and 2017 [Citation109]. In this study, fewer than 5% of patients had a severe adverse event, and success rates of FMT increased over time, exceeding 75% in the last several years of the study [Citation109]. With the success of FMT in treating children with recurrent CDI, use of this procedure has more recently expanded to include other pediatric patient populations and indications, with small case series demonstrating possible benefits of FMT in children being evaluated for HCT who are colonized by multidrug-resistant organisms, children with inflammatory bowel disease, and children with severe acute graft-versus-host disease following allogeneic HCT [Citation110–112]. FMT has also been studied as an approach to modify the gut microbiota of infants born via C-section delivery. In one small study involving seven mother-to-infant FMTs, the gut microbiomes of infants born via C-section who were treated with maternal FMT were more similar to the gut microbiomes of vaginally delivered infants than those of infants born by C-section who did not undergo FMT [Citation113]. With the accumulating data regarding the safety of FMT in children, it is likely that use of this procedure will expand in pediatric patient populations in the coming years.
3.3. Phage therapy
Phage therapy, in which viruses that target and kill bacteria (bacteriophages) are used for infection treatment, dates back more than a century; however, there has been renewed interest in phages as a result of rising antibiotic resistance. Few controlled trials of phage therapy have been conducted among children; most notably, a clinical trial of Escherichia coli bacteriophages in children presenting with acute diarrhea was stopped early due to a lack of clinical and microbiological response [Citation114]. However, there have been several reports of treatment of children with multidrug-resistant bacterial infections with phage therapy that have reported favorable clinical outcomes [Citation115–117]. Similarly, in a case series of phage therapy for nontuberculous mycobacterial infections that included children and adults, a favorable clinical or microbiological response was observed in 11 of 20 patients, with the varied treatment responses possibly being related to heterogeneity of host risk factors, characteristics of the infections, and the poorly understood pharmacodynamics of phage therapy [Citation118]. Several controlled studies of phage therapy are ongoing, including a study that will evaluate the efficacy of phage therapy in children and adults who have exhausted other treatment options for infections [Citation119]. With the global rise in antibiotic resistance and significant gaps in the preclinical and clinical antibiotic pipelines, it is likely that phage therapy will continue to be explored as a treatment option for children with infections caused by multidrug-resistant bacteria.
4. The gut microbiota and immune responses to childhood vaccination
Vaccines have contributed to an unprecedented reduction in child mortality over the past century and continue to prevent nearly 3 million child deaths each year [Citation120]. However, for reasons that remain poorly understood, the immunogenicity and effectiveness of vaccines are highly variable between children within the same population and across different populations of children. For instance, oral rotavirus vaccines are less immunogenic and less effective in preventing severe rotavirus-associated diarrhea among children in low- and middle-income countries than among children in high-income countries [Citation121–124]. Although several factors are likely to contribute to these individual and population-level differences in vaccine-elicited immunity, the gut microbiota is increasingly recognized to play a significant role in modulating development of these immune responses.
Perhaps the most compelling data supporting an association between the gut microbiota and responses to childhood vaccines comes from experiments conducted in animal models. In a landmark study, Lynn and colleagues demonstrated that infant mice that were exposed to ampicillin and neomycin prior to receipt of inactivated or live vaccines developed inferior vaccine-induced antibody responses [Citation125]. Moreover, rescue of antibiotic-treated mice with FMT from untreated mice prior to administration of 13-valent pneumococcal conjugate vaccine restored antibody responses to this vaccine, whereas antibody responses remained inferior among antibiotic-treated mice that received FMT from other antibiotic-treated mice [Citation125]. Interestingly, treatment of adult mice with antibiotics did not impair humoral immune responses to pneumococcal conjugate vaccine, suggesting that vaccine-elicited immune responses may be particularly susceptible to modulation by the gut microbiota early in life [Citation125]. Other studies conducted using animal models have similarly shown that the gut microbiota can influence immune responses to vaccines for hepatitis B virus, rabies virus, Mycobacterium tuberculosis, and avian influenza virus [Citation126–129].
Findings from clinical studies of the impact of the gut microbiota on vaccine-induced immunity have demonstrated varied and often conflicting results (). Several correlative clinical studies reported associations between gut microbiota composition and the response to specific childhood vaccines. For instance, Harris and colleagues found that the gut microbiota composition of infants in Ghana administered oral rotavirus vaccine differed based on vaccine response, with vaccine responders having a higher abundance of Streptococcus bovis and a lower abundance of the bacterial phylum Bacteroidetes than nonresponders [Citation132]. Further, the gut microbiota of Ghanaian vaccine responders was actually more similar to that of Dutch infants than to Ghanaian vaccine nonresponders [Citation132], suggesting that gut microbiota composition may contribute to the varied effectiveness of oral rotavirus vaccines in low- and middle-income compared to high-income countries. Huda and colleagues similarly reported that higher gut abundances of the bacterial phylum Actinobacteria were associated with more robust cellular immune responses to oral poliovirus, bacille Calmette-Guérin (BCG), and hepatitis B virus vaccines, while the abundance of Rothia correlated positively with antibody responses to tetanus toxoid [Citation136]. Notably, the gut microbes identified as being associated with vaccine responses have generally not been consistent across clinical studies, while other such correlative studies have reported no associations between the gut microbiota and vaccine responses [Citation130,Citation131,Citation134,Citation135]. These varied findings across studies could reflect the small sample sizes of these studies, biological differences in the populations being studied, or residual confounding by nutritional, socioeconomic, or environmental factors.
Table 1. Correlative clinical studies that evaluated the bacterial gut microbiome as a modulator of immune responses to vaccination.
With the recognition that the gut microbiota may influence immune responses to vaccination comes the possibility that targeted modification of the gut microbiota could promote vaccine-elicited immunity. In particular, there has been substantial interest in the use of probiotics as vaccine adjuvants. Isolauri and colleagues reported that oral administration of Lactobacillus casei strain GG was associated with a more robust serum antibody response to oral rotavirus vaccine than placebo among 54 infants in Finland [Citation138]. Similarly, Kukkonen and colleagues found that a consortium of four bacterial strains was associated with the development of higher quantitative antibody responses to Haemophilus influenzae type B vaccine than placebo among Finnish infants, although antibody responses to diphtheria and tetanus did not differ in these groups [Citation139]. While promising, findings from these studies have not been evaluated in large clinical trials, and probiotics did not improve vaccine immunogenicity in several other trials conducted among young children [Citation140–142]. Unfortunately, the substantial variability in the composition and timing of administration of the probiotics evaluated in the clinical trials conducted to date precludes comparisons of findings across studies. Finally, a single study evaluated the impact of antibiotic treatment on vaccine-elicited immune responses among children. In a placebo-controlled trial that included 754 Indian infants, treatment with azithromycin before vaccination did not improve the immunogenicity of oral poliovirus vaccine despite reducing fecal biomarkers of environmental enteropathy and the prevalence of enteric pathogens [Citation143].
5. Conclusion
In conclusion, the human microbiota undergoes marked shifts in composition during early life as a result of environmental exposures. During infancy, these microbes are essential to immune maturation, promoting the generation of protective immune responses to potential pathogens and the development of immune tolerance to commensal microbes and host antigens. Throughout childhood, the commensal microbiota influences the risk and severity of infections through a complex network of microbe–microbe and microbe–host interactions collectively referred to as colonization resistance. This important function of the microbiota influences the risk of many infectious diseases that are common among healthy children and adolescents, including acute respiratory infections, enteric infections, and sexually transmitted infections. Moreover, the microbiota plays a key role in the pathogenesis of gut-derived BSIs among vulnerable groups of children, including premature infants and pediatric HCT recipients. Data from animal experiments and correlative human studies also suggest that the microbiota influences immune responses to childhood vaccines. Taken together, the studies highlighted in this review demonstrate the substantial influence of the microbiota on infectious diseases during childhood and the enormous potential of microbiome-based therapies to prevent or treat infections and promote vaccine-elicited immunity among children.
6. Expert opinion
Our understanding of the influence of the microbiota on childhood infections has advanced remarkably over the past several decades. However, several important gaps in knowledge remain. To date, we know surprisingly little regarding factors that shape microbial communities and their influence on infections outside of the gastrointestinal and respiratory tracts or in older children and adolescents. For instance, few prior studies have investigated the microbiota of the male reproductive tract and the extent to which it influences the transmission of STIs among adolescents. Additionally, the overwhelming majority of studies of the early life microbiome have focused solely on bacteria, effectively disregarding the potential impact of microbes from other kingdoms on childhood infections. Finally, there is growing recognition that the microbiome field needs to move beyond correlative studies in order to accelerate the development of microbiome-based interventions to prevent or treat infections and other diseases in children.
One of the main challenges facing the field is translating what we have learned about the microbiome to develop effective diagnostics and therapeutics for childhood infections. Next-generation sequencing technologies have the potential to revolutionize our approach to diagnosing infections. Recently, several commercial laboratories have developed assays that enable detection of clinically relevant microbes in a variety of human samples with a short turnaround time, providing results that can be used to guide clinical decision-making. These sequencing technologies also have the potential to transform the field of infection prevention and control. Multilocus sequence typing, which involves sequencing of only a handful of microbial genes that typically represent less than 1% of the genome, remains a standard approach for the evaluation of healthcare-associated infection clusters [Citation144]. The increasing use of whole-genome sequencing, which enables analyzes of entire genomes and has far higher resolution for strain comparisons than multilocus sequence typing, has the potential to improve allocation of the limited resources, time, and personnel available for infection prevention and control investigations [Citation145].
Since November 2022, the first two microbiota-based human therapeutics for adults with recurrent C. difficile infection have been approved by the United States Food and Drug Administration (FDA). While the approval of these products represents a major milestone for the microbiome field, there are still no FDA-approved microbiota-based therapies for the prevention or treatment of other infections or for use in children. One potential barrier to the development of microbiome-based therapies relates to the current regulatory approaches to therapeutic approval. Many microbiome-based therapies developed for infection prevention are likely to act by blocking or eradicating pathogen colonization as a precursor to infection. Colonization by potential pathogens is most frequently an asymptomatic state that would fall outside of the outcomes that have historically been used for therapeutic approval by regulatory bodies. Moreover, the sample sizes that would be needed to demonstrate that such therapies reduce the incidence of symptomatic infections may be prohibitively large. To realize the full potential of microbiome-based therapies for infection prevention, regulatory approval processes may need to be modified to consider pathogen colonization as a proxy for infection and as a valid clinical trial outcome for product approval. Another challenge to the development of microbiome-based therapies for children is that pediatric populations are often excluded from clinical trials until therapeutics have been shown to be effective in adults. As described in this review, the microbiomes of children differ markedly from those of adults, and early life may represent a critical period during which the microbiome is more malleable to microbiome-targeted interventions. Thus, some microbiome-based therapies may only have an effect in children or may be more effective in children than in adults, which could lead to such products being abandoned if pediatric populations are not included in early clinical trials.
As sequencing technologies and analytic methods become increasingly high-throughput and accessible, we anticipate that the study of the child microbiome will advance in several ways over the next 5 years. First, there will be an increase in studies investigating the extent to which fungi, viruses, archaea, and protozoa influence childhood infections through both microbe-host and microbe–microbe interactions, including interspecies interactions spanning microbial kingdoms. Moreover, we will see a continued shift from simple correlative studies to translational studies that utilize novel experimental models or that integrate data on microbial gene expression, microbial and host metabolites, and host immune responses. Additionally, commercial next-generation sequencing assays will be further validated as diagnostic tools and incorporated into routine clinical practice. Eventually, in time, the pathways for regulatory review and approval will be updated to enable development of an increasingly broad range of microbiome-based therapies that reduce the burden of infectious diseases among children.
Article highlights
The microbiome resists colonization and infection by exogenous pathogens through diverse mechanisms collectively referred to as colonization resistance.
Shifts in microbiome composition during early childhood impact immune maturation and can have lasting effects on infection susceptibility and severity.
Microbiome-based therapies have the potential to prevent and treat infectious diseases during childhood.
The gut microbiota may impact the immunogenicity of routine childhood vaccinations.
Declaration of interest
MS Kelly receives grant funding from and serves as a consultant for Merck & Co., Inc. MS Kelly serves on an advisory board for Invivyd.
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Additional information
Funding
References
- Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016 Aug;14(8):e1002533. doi: 10.1371/journal.pbio.1002533
- Tierney BT, Yang Z, Luber JM, et al. The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe. 2019;26(2):283–295. e8. doi: 10.1016/j.chom.2019.07.008
- Beller L, Deboutte W, Falony G, et al. Successional stages in infant gut microbiota maturation. MBio. 2021 Dec 21;12(6):e0185721. doi: 10.1128/mbio.01857-21
- Arrieta MC, Stiemsma LT, Amenyogbe N, et al. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427
- Xiao L, Wang J, Zheng J, et al. Deterministic transition of enterotypes shapes the infant gut microbiome at an early age. Genome Biol. 2021 Aug 24;22(1):243. doi: 10.1186/s13059-021-02463-3
- Lopera-Maya EA, Kurilshikov A, van der Graaf A, et al. Effect of host genetics on the gut microbiome in 7,738 participants of the Dutch microbiome project. Nat Genet. 2022 Feb;54(2):143–151.
- Kurilshikov A, Medina-Gomez C, Bacigalupe R, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021 Feb;53(2):156–165.
- Shao Y, Forster SC, Tsaliki E, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019 Oct;574(7776):117–121.
- Bosch A, Levin E, van Houten MA, et al. Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine. 2016 Jul;9:336–345. doi: 10.1016/j.ebiom.2016.05.031
- Chu DM, Ma J, Prince AL, et al. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017 Mar;23(3):314–326.
- Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010 Jun 29;107(26):11971–11975. doi: 10.1073/pnas.1002601107
- Barnett DJM, Endika MF, Klostermann CE, et al. Human milk oligosaccharides, antimicrobial drugs, and the gut microbiota of term neonates: observations from the KOALA birth cohort study. Gut Microbes. 2023 Jan;15(1):2164152.
- Holst AQ, Myers P, Rodriguez-Garcia P, et al. Infant formula supplemented with five human milk oligosaccharides shifts the fecal microbiome of formula-fed infants closer to that of breastfed infants. Nutrients. 2023 Jul 10;15(14):3087. doi: 10.3390/nu15143087
- Reyman M, van Houten MA, Watson RL, et al. Effects of early-life antibiotics on the developing infant gut microbiome and resistome: a randomized trial. Nat Commun. 2022 Feb 16;13(1):893. doi: 10.1038/s41467-022-28525-z
- Claassen-Weitz S, Gardner-Lubbe S, Xia Y, et al. Succession and determinants of the early life nasopharyngeal microbiota in a South African birth cohort. Microbiome. 2023 Jun 5;11(1):127. doi: 10.1186/s40168-023-01563-5
- Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012 May 9;486(7402):222–227. doi: 10.1038/nature11053
- Hollister EB, Riehle K, Luna RA, et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015 Aug 26;3(1):36. doi: 10.1186/s40168-015-0101-x
- Raman AS, Gehrig JL, Venkatesh S, et al. A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science. 2019 Jul 12;365(6449). doi: 10.1126/science.aau4735
- Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science (New York, NY). Feb 1. 2013;339(6119):548–554. doi: 10.1126/science.1229000
- Gehrig JL, Venkatesh S, Chang HW, et al. Effects of microbiota-directed foods in gnotobiotic animals and undernourished children. Science. 2019 Jul 12;365(6449). doi: 10.1126/science.aau4732
- Korpela K, Salonen A, Virta LJ, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. 2016 Jan 26;7(1):10410. doi: 10.1038/ncomms10410
- Abushawish A, Haro K, Hoshina T, et al. Environmental factors related to differences in the microbiota in the upper respiratory tract in young children: Focusing on the impact of early nursery attendance. Front Pediatr. 2023;11:1015872. doi: 10.3389/fped.2023.1015872
- Amir A, Erez-Granat O, Braun T, et al. Gut microbiome development in early childhood is affected by day care attendance. NPJ Biofilms Microbiomes. 2022 Jan 11;8(1):2. doi: 10.1038/s41522-021-00265-w
- Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018 Oct;562(7728):583–588.
- Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. 1954 May;86(1):132–137. doi: 10.3181/00379727-86-21030
- Osbelt L, Wende M, Almasi E, et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe. 2021 Nov 10;29(11):1663–1679 e7. doi: 10.1016/j.chom.2021.09.003
- Robertson RC, Manges AR, Finlay BB, et al. The human microbiome and child growth - first 1000 days and beyond. Trends Microbiol. 2019 Feb;27(2):131–147.
- Gensollen T, Iyer SS, Kasper DL, et al. How colonization by microbiota in early life shapes the immune system. Science. 2016 Apr 29;352(6285):539–544. doi: 10.1126/science.aad9378
- Torow N, Hornef MW. The neonatal window of opportunity: Setting the stage for life-long host-microbial interaction and immune homeostasis. J Immunol. 2017 Jan 15;198(2):557–563. doi: 10.4049/jimmunol.1601253
- Abrahamsson TR, Jakobsson HE, Andersson AF, et al. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014 Jun;44(6):842–850.
- Marra F, Marra CA, Richardson K, et al. Antibiotic use in children is associated with increased risk of asthma. Pediatrics. 2009 Mar;123(3):1003–1010.
- Boutin RCT, Sbihi H, Dsouza M, et al. Mining the infant gut microbiota for therapeutic targets against atopic disease. Allergy. 2020 Aug;75(8):2065–2068.
- Arildsen AW, Zachariassen LF, Krych L, et al. Delayed gut colonization shapes future allergic responses in a murine model of atopic dermatitis. Front Immunol. 2021;12:650621. doi: 10.3389/fimmu.2021.650621
- Basha S, Surendran N, Pichichero M. Immune responses in neonates. Expert Rev Clin Immunol. 2014 Sep;10(9):1171–1184. doi: 10.1586/1744666X.2014.942288
- Neish AS. Mucosal immunity and the microbiome. Ann Am Thorac Soc. 2014 Jan;11(Suppl 1):S28–32. doi: 10.1513/AnnalsATS.201306-161MG
- Ciabattini A, Olivieri R, Lazzeri E, et al. Role of the microbiota in the modulation of vaccine immune responses. Front Microbiol. 2019;10:1305. doi: 10.3389/fmicb.2019.01305
- Chiu CY, Chan YL, Tsai MH, et al. Gut microbial dysbiosis is associated with allergen-specific IgE responses in young children with airway allergies. World Allergy Organ J. 2019;12(3):100021. doi: 10.1016/j.waojou.2019.100021
- Bullard BM, VanderVeen BN, McDonald SJ, et al. Cross talk between the gut microbiome and host immune response in ulcerative colitis: nonpharmacological strategies to improve homeostasis. Am J Physiol Gastrointest Liver Physiol. 2022 Dec 1;323(6):G554–G561. doi: 10.1152/ajpgi.00210.2022
- Zheng L, Kelly CJ, Battista KD, et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor–dependent repression of Claudin-2. J Immunol. 2017 Oct 15;199(8):2976–2984. doi: 10.4049/jimmunol.1700105
- Correa-Oliveira R, Fachi JL, Vieira A, et al. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016 Apr;5(4):e73.
- Vaishnava S, Yamamoto M, Severson KM, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011 Oct 14;334(6053):255–258. doi: 10.1126/science.1209791
- Brandl K, Plitas G, Mihu CN, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008 Oct 9;455(7214):804–807. doi: 10.1038/nature07250
- Sequeira RP, McDonald JAK, Marchesi JR, et al. Commensal Bacteroidetes protect against Klebsiella pneumoniae colonization and transmission through IL-36 signalling. Nat Microbiol. 2020 Feb;5(2):304–313.
- Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun. 2017 Nov 15;8(1):1512. doi: 10.1038/s41467-017-01803-x
- Safiri S, Mahmoodpoor A, Kolahi AA, et al. Global burden of lower respiratory infections during the last three decades. Front Public Health. 2022;10:1028525. doi: 10.3389/fpubh.2022.1028525
- Kelly MS, Plunkett C, Yu Y, et al. Non-diphtheriae Corynebacterium species are associated with decreased risk of pneumococcal colonization during infancy. Isme J. 2022 Mar;16(3):655–665.
- Stubbendieck RM, Dissanayake E, Burnham PM, et al. Rothia from the human nose inhibit Moraxella catarrhalis Colonization with a secreted Peptidoglycan Endopeptidase. MBio. 2023 Apr 25;14(2):e0046423. doi: 10.1128/mbio.00464-23
- Deasy AM, Guccione E, Dale AP, et al. Nasal inoculation of the commensal neisseria lactamica inhibits carriage of Neisseria meningitidis by young adults: A controlled human infection study. Clin Infect Dis. 2015 May 15;60(10):1512–1520. doi: 10.1093/cid/civ098
- Uehara Y, Nakama H, Agematsu K, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect. 2000 Feb;44(2):127–133.
- de Steenhuijsen Piters WAA, Watson RL, de Koff EM, et al. Early-life viral infections are associated with disadvantageous immune and microbiota profiles and recurrent respiratory infections. Nat Microbiol. 2022 Feb;7(2):224–237.
- Dissanayake E, Brockman-Schneider RA, Stubbendieck RM, et al. Rhinovirus increases Moraxella catarrhalis adhesion to the respiratory epithelium. Front Cell Infect Microbiol. 2022;12:1060748. doi: 10.3389/fcimb.2022.1060748
- Gu L, Deng H, Ren Z, et al. Dynamic changes in the microbiome and mucosal immune microenvironment of the lower respiratory tract by influenza virus infection. Front Microbiol. 2019;10:2491. doi: 10.3389/fmicb.2019.02491
- Hurst JH, McCumber AW, Aquino JN, et al. Age-related changes in the Nasopharyngeal Microbiome are associated with severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) infection and symptoms among children, adolescents, and young adults. Clin Infect Dis. 2022 Aug 24;75(1):e928–e937. doi: 10.1093/cid/ciac184
- de Steenhuijsen Piters WA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016 Nov 1;194(9):1104–1115. doi: 10.1164/rccm.201602-0220OC
- Koenen MH, de Groot RCA, de Steenhuijsen Piters WAA, et al. Mycoplasma pneumoniae carriage in children with recurrent respiratory tract infections is associated with a less diverse and altered microbiota. EBioMedicine. 2023 Nov 9;98:104868. doi: 10.1016/j.ebiom.2023.104868
- Penela-Sanchez D, Rocafort M, Henares D, et al. Impact of the bacterial nasopharyngeal microbiota on the severity of genus enterovirus lower respiratory tract infection in children: a case–control study. Pediatr Pulmonol. 2023 Jun;58(6):1728–1737.
- Ling Z, Liu X, Jia X, et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep. 2014 Dec 15;4(1):7485. doi: 10.1038/srep07485
- Schubert AM, Rogers MA, Ring C, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014 May 6;5(3):e01021–14. doi: 10.1128/mBio.01021-14
- Berkell M, Mysara M, Xavier BB, et al. Microbiota-based markers predictive of development of clostridioides difficile infection. Nat Commun. 2021 Apr 14;12(1):2241. doi: 10.1038/s41467-021-22302-0
- Smith AB, Jenior ML, Keenan O, et al. Enterococci enhance clostridioides difficile pathogenesis. Nature. 2022 Nov;611(7937):780–786.
- Brennhofer SA, Rogawski McQuade ET, Liu J, et al. Clostridioides difficile colonization among very young children in resource-limited settings. Clin Microbiol Infect. 2022 Jul;28(7):996–1002.
- Mani J, Levy S, Angelova A, et al. Epidemiological and microbiome associations of Clostridioides difficile carriage in infancy and early childhood. Gut Microbes. 2023 Jan;15(1):2203969.
- Kociolek LK, Espinosa RO, Gerding DN, et al. Natural clostridioides difficile toxin immunization in colonized infants. Clin Infect Dis. 2020 May 6;70(10):2095–2102. doi: 10.1093/cid/ciz582
- Couturier J, Lepage P, Jolivet S, et al. Gut microbiota diversity of preterm neonates is associated with clostridioides difficile colonization. Front Cell Infect Microbiol. 2022;12:907323. doi: 10.3389/fcimb.2022.907323
- Singer JR, Blosser EG, Zindl CL, et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat Med. 2019 Nov;25(11):1772–1782.
- Heston SM, Lim CSE, Ong C, et al. Strain-resolved metagenomic analysis of the gut as a reservoir for bloodstream infection pathogens among premature infants in Singapore. Gut Pathog. 2023;15(1):55. doi: 10.1186/s13099-023-00583-8
- Kelly MS, Ward DV, Severyn CJ, et al. Gut colonization preceding mucosal barrier injury bloodstream infection in pediatric hematopoietic stem cell transplantation recipients. Biol Blood Marrow Transplant. 2019 Nov;25(11):2274–2280.
- Margolis EB, Alfaro GM, Sun Y, et al. Microbiota predict infections and acute graft-versus-host disease after pediatric allogeneic hematopoietic stem cell transplantation. J Infect Dis. 2023 May 30;228(5):627–636. doi: 10.1093/infdis/jiad190
- Bargheet A, Klingenberg C, Esaiassen E, et al. Development of early life gut resistome and mobilome across gestational ages and microbiota-modifying treatments. EBioMedicine. 2023 Jun;92:104613. doi: 10.1016/j.ebiom.2023.104613
- Moore AM, Patel S, Forsberg KJ, et al. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLOS ONE. 2013;8(11):e78822. doi: 10.1371/journal.pone.0078822
- Carvalho MJ, Sands K, Thomson K, et al. Antibiotic resistance genes in the gut microbiota of mothers and linked neonates with or without sepsis from low- and middle-income countries. Nat Microbiol. 2022 Sep;7(9):1337–1347.
- Pereira-Dias J, Nguyen Ngoc Minh C, Tran Thi Hong C, et al. The gut microbiome of healthy vietnamese adults and children is a major reservoir for resistance genes against critical antimicrobials. J Infect Dis. 2021 Dec 20;224(12 Suppl 2):S840–S847. doi: 10.1093/infdis/jiab398
- Doan T, Worden L, Hinterwirth A, et al. Macrolide and nonmacrolide resistance with mass Azithromycin distribution. N Engl J Med. 2020 Nov 12;383(20):1941–1950. doi: 10.1056/NEJMoa2002606
- Gibson MK, Wang B, Ahmadi S, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. 2016 Mar 7;1(4):16024. doi: 10.1038/nmicrobiol.2016.24
- Centers for Disease Control and Prevention. Sexually transmited diseases surveillance 2021. Available at: https://www.cdc.gov/std/statistics/2021/default.htm. Accessed Dec 15, 2023.
- Centers for Disease Control and Prevention. Estimated HIV incidence and prevalence in the United States, 2017-2021. cited 2023 Dec 15. Available from: https://www.cdc.gov/hiv/library/reports/hiv-surveillance/vol-28-no-3/index.html
- Sexually transmitted disease surveillance 2021 cdc.Gov: center for disease control and prevention; 2023 [updated April 11, 2023; cited 2023 November 20]. Available from: https://www.cdc.gov/std/statistics/2021/tables.htm
- Chen X, Lu Y, Chen T, et al. The female vaginal microbiome in health and bacterial vaginosis. Front Cell Infect Microbiol. 2021;11:631972. doi: 10.3389/fcimb.2021.631972
- Witkin SS, Linhares IM. Why do lactobacilli dominate the human vaginal microbiota? BJOG: An Int J Obstetrics Gynaecol. 2017;124(4):606–611. doi: 10.1111/1471-0528.14390
- Tuddenham S, Ravel J, Marrazzo JM. Protection and risk: Male and female genital microbiota and sexually transmitted infections. J Infect Dis. 2021 Jun 16;223(12 Suppl 2):S222–S235. doi: 10.1093/infdis/jiaa762
- Sena AC, Goldstein LA, Ramirez G, et al. Bacterial vaginosis and its association with incident Trichomonas vaginalis infections: A systematic review and meta-analysis. Sex Transm Dis. 2021 Dec 1;48(12):e192–e201. doi: 10.1097/OLQ.0000000000001537
- Tamarelle J, Thiebaut ACM, de Barbeyrac B, et al. The vaginal microbiota and its association with human papillomavirus, Chlamydia trachomatis, Neisseria gonorrhoeae and Mycoplasma genitalium infections: a systematic review and meta-analysis. Clin Microbiol Infect. 2019 Jan;25(1):35–47.
- Gholizadeh O, Yekanipour Z, Kivi S, et al. Review of the evidence of the effects of human papillomavirus infection and Gardnerella vaginalis, and their co-infection on infertility. Microb Pathog. 2023 Feb;175:105980. doi: 10.1016/j.micpath.2023.105980
- Nicolo S, Tanturli M, Mattiuz G, et al. Vaginal lactobacilli and vaginal dysbiosis-associated bacteria differently affect cervical epithelial and immune homeostasis and anti-viral defenses. Int J Mol Sci. 2021 Jun 17;22(12):6487. doi: 10.3390/ijms22126487
- Lovett A, Sena AC, Macintyre AN, et al. Cervicovaginal microbiota predicts neisseria gonorrhoeae clinical presentation. Front Microbiol. 2021;12:790531. doi: 10.3389/fmicb.2021.790531
- Atashili J, Poole C, Ndumbe PM, et al. Bacterial vaginosis and HIV acquisition: a meta-analysis of published studies. AIDS. 2008 Jul 31;22(12):1493–1501. doi: 10.1097/QAD.0b013e3283021a37
- McClelland RS, Lingappa JR, Srinivasan S, et al. Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study. Lancet Infect Dis. 2018 May;18(5):554–564.
- Karim SS, Kashuba AD, Werner L, et al. Drug concentrations after topical and oral antiretroviral pre-exposure prophylaxis: implications for HIV prevention in women. Lancet. 2011 Jul 16;378(9787):279–281. doi: 10.1016/S0140-6736(11)60878-7
- Grant RM, Lama JR, Anderson PL, et al. Preexposure chemoprophylaxis for HIV prevention in men who have sex with men. N Engl J Med. 2010 Dec 30;363(27):2587–2599. doi: 10.1056/NEJMoa1011205
- McCormack S, Dunn DT, Desai M, et al. Pre-exposure prophylaxis to prevent the acquisition of HIV-1 infection (PROUD): effectiveness results from the pilot phase of a pragmatic open-label randomised trial. Lancet. 2016 Jan 2;387(10013):53–60. doi: 10.1016/S0140-6736(15)00056-2
- Molina JM, Capitant C, Spire B, et al. On-demand preexposure prophylaxis in men at high risk for HIV-1 infection. N Engl J Med. 2015 Dec 3;373(23):2237–2246. doi: 10.1056/NEJMoa1506273
- Klatt NR, Cheu R, Birse K, et al. Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science. 2017 Jun 2;356(6341):938–945. doi: 10.1126/science.aai9383
- Dashper SG, Mitchell HL, Le Cao KA, et al. Temporal development of the oral microbiome and prediction of early childhood caries. Sci Rep. 2019 Dec 24;9(1):19732. doi: 10.1038/s41598-019-56233-0
- Kim D, Barraza JP, Arthur RA, et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci U S A. 2020 Jun 2;117(22):12375–12386. doi: 10.1073/pnas.1919099117
- Okada M, Kawamura M, Oda Y, et al. Caries prevalence associated with Streptococcus mutans and Streptococcus sobrinus in Japanese schoolchildren. Int J Paediatr Dent. 2012 Sep;22(5):342–348.
- Havsed K, Stensson M, Jansson H, et al. Bacterial composition and metabolomics of dental plaque from adolescents. Front Cell Infect Microbiol. 2021;11:716493. doi: 10.3389/fcimb.2021.716493
- Teng F, Yang F, Huang S, et al. Prediction oF early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe. 2015 Sep 9;18(3):296–306. doi: 10.1016/j.chom.2015.08.005
- Wang Y, Florez ID, Morgan RL, et al. Probiotics, prebiotics, lactoferrin, and combination products for prevention of mortality and morbidity in preterm infants: A systematic review and network meta-analysis. JAMA Pediatr. 2023 Nov 1;177(11):1158–1167. doi: 10.1001/jamapediatrics.2023.3849
- Beck LC, Masi AC, Young GR, et al. Strain-specific impacts of probiotics are a significant driver of gut microbiome development in very preterm infants. Nat Microbiol. 2022 Oct;7(10):1525–1535.
- Samara J, Moossavi S, Alshaikh B, et al. Supplementation with a probiotic mixture accelerates gut microbiome maturation and reduces intestinal inflammation in extremely preterm infants. Cell Host Microbe. 2022 May 11;30(5):696–711.e5. doi: 10.1016/j.chom.2022.04.005
- Jacobsen CN, Rosenfeldt Nielsen V, Hayford A, et al. Screening of probiotic activities of forty-seven strains of lactobacillus spp. By in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl environ microbiol. 1999;65(11):4949–4956. doi: 10.1128/AEM.65.11.4949-4956.1999
- Kristensen NB, Bryrup T, Allin KH, et al. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8(1):1–11. doi: 10.1186/s13073-016-0300-5
- Guitor AK, Yousuf EI, Raphenya AR, et al. Capturing the antibiotic resistome of preterm infants reveals new benefits of probiotic supplementation. Microbiome. 2022 Aug 26;10(1):136. doi: 10.1186/s40168-022-01327-7
- Casaburi G, Duar RM, Vance DP, et al. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob Resist Infect Control. 2019;8(1):131. doi: 10.1186/s13756-019-0583-6
- Mourey F, Sureja V, Kheni D, et al. A multicenter, randomized, double-blind, placebo-controlled trial of saccharomyces boulardii in infants and children with Acute Diarrhea. Pediatr Infect Dis J. 2020 Nov;39(11):e347–e351.
- Horne RG, Freedman SB, Johnson-Henry KC, et al. Intestinal microbial composition of children in a randomized controlled trial of probiotics to treat acute gastroenteritis. Front Cell Infect Microbiol. 2022;12:883163. doi: 10.3389/fcimb.2022.883163
- Guo Q, Goldenberg JZ, Humphrey C, et al. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. 2019 Apr 30;4(4):Cd004827. doi: 10.1002/14651858.CD004827.pub5
- Goldenberg JZ, Yap C, Lytvyn L, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2017 Dec 19;12(12):Cd006095. doi: 10.1002/14651858.CD006095.pub4
- Nicholson MR, Mitchell PD, Alexander E, et al. Efficacy of fecal microbiota transplantation for Clostridium difficile infection in children. Clin Gastroenterol Hepatol. 2020;18(3):612–619. e1. doi: 10.1016/j.cgh.2019.04.037
- Merli P, Putignani L, Ruggeri A, et al. Decolonization of multi-drug resistant bacteria by fecal microbiota transplantation in five pediatric patients before allogeneic hematopoietic stem cell transplantation: gut microbiota profiling, infectious and clinical outcomes. Haematologica. 2020;105(11):2686. doi: 10.3324/haematol.2019.244210
- Goyal A, Yeh A, Bush BR, et al. Safety, clinical response, and microbiome findings following fecal microbiota transplant in children with inflammatory bowel disease. Inflamm Bowel Dis. 2018;24(2):410–421. doi: 10.1093/ibd/izx035
- Goloshchapov OV, Bakin EA, Stanevich OV, et al. Clinical and immune effects of fecal microbiota transplantation in children with acute graft-versus-host disease. Cellular Therapy And Transplant. 2021;10(1):69–78. doi: 10.18620/ctt-1866-8836-2021-10-1-69-78
- Korpela K, Helve O, Kolho KL, et al. Maternal fecal microbiota transplantation in cesarean-born infants rapidly restores normal gut microbial development: A proof-of-concept study. Cell. 2020 Oct 15;183(2):324–334.e5. doi: 10.1016/j.cell.2020.08.047
- Sarker SA, Sultana S, Reuteler G, et al. Oral phage therapy of acute bacterial diarrhea with two coliphage preparations: a randomized trial in children from Bangladesh. EBioMedicine. 2016 Feb;4:124–137. doi: 10.1016/j.ebiom.2015.12.023
- Duplessis C, Biswas B, Hanisch B, et al. Refractory Pseudomonas Bacteremia in a 2-year-old sterilized by bacteriophage therapy. J Pediatric Infect Dis Soc. 2018 Aug 17;7(3):253–256. doi: 10.1093/jpids/pix056
- Paul K, Merabishvili M, Hazan R, et al. Bacteriophage rescue therapy of a vancomycin-resistant enterococcus faecium infection in a one-year-old child following a third liver transplantation. Viruses. 2021 Sep 7;13(9):1785. doi: 10.3390/v13091785
- Bradley JS, Hajama H, Akong K, et al. Bacteriophage therapy of multidrug-resistant achromobacter in an 11-year-old boy with cystic fibrosis assessed by metagenome analysis. Pediatr Infect Dis J. 2023 Sep 1;42(9):754–759. doi: 10.1097/INF.0000000000004000
- Dedrick RM, Smith BE, Cristinziano M, et al. Phage therapy of mycobacterium infections: Compassionate use of phages in 20 patients with drug-resistant mycobacterial disease. Clin Infect Dis. 2023 Jan 6;76(1):103–112. doi: 10.1093/cid/ciac453
- Khatami A, Foley DA, Warner MS, et al. Standardised treatment and monitoring protocol to assess safety and tolerability of bacteriophage therapy for adult and paediatric patients (STAMP study): protocol for an open-label, single-arm trial. BMJ Open. 2022 Dec 9;12(12):e065401. doi: 10.1136/bmjopen-2022-065401
- Secretary-General U Progress at mid-decade on implementation of general assembly resolution 45/217 on the world summit for children: report of the secretary-general. 1996.
- Armah GE, Sow SO, Breiman RF, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376(9741):606–614. doi: 10.1016/S0140-6736(10)60889-6
- Vesikari T, Karvonen A, Prymula R, et al. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet. 2007;370(9601):1757–1763. doi: 10.1016/S0140-6736(07)61744-9
- Zaman K, Anh DD, Victor JC, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;376(9741):615–623. doi: 10.1016/S0140-6736(10)60755-6
- Madhi SA, Cunliffe NA, Steele D, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. N Engl J Med. 2010;362(4):289–298. doi: 10.1056/NEJMoa0904797
- Lynn MA, Tumes DJ, Choo JM, et al. Early-life antibiotic-driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe. 2018;23(5):653–660. e5. doi: 10.1016/j.chom.2018.04.009
- Swaminathan G, Citron M, Xiao J, et al. Vaccine hyporesponse induced by individual antibiotic treatment in mice and non-human primates is diminished upon recovery of the gut microbiome. Vaccines. 2021;9(11):1340. doi: 10.3390/vaccines9111340
- Yitbarek A, Astill J, Hodgins DC, et al. Commensal gut microbiota can modulate adaptive immune responses in chickens vaccinated with whole inactivated avian influenza virus subtype H9N2. Vaccine. 2019;37(44):6640–6647. doi: 10.1016/j.vaccine.2019.09.046
- Nadeem S, Maurya SK, Das DK, et al. Gut dysbiosis thwarts the efficacy of vaccine against Mycobacterium tuberculosis. Front Immunol. 2020;11:726. doi: 10.3389/fimmu.2020.00726
- Zhang Y, Wu Q, Zhou M, et al. Composition of the murine gut microbiome impacts humoral immunity induced by rabies vaccines. Clin Transl Med. 2020;10(4):e161. doi: 10.1002/ctm2.161
- Praharaj I, Parker EPK, Giri S, et al. Influence of nonpolio enteroviruses and the bacterial gut microbiota on oral poliovirus vaccine response: a study from South India. J Infect Dis. 2018;219(8):1178–1186. doi: 10.1093/infdis/jiy568
- Zhao T, Li J, Fu Y, et al. Influence of gut microbiota on mucosal IgA antibody response to the polio vaccine. NPJ Vaccin. 2020 Jun 09;5(1):47. doi: 10.1038/s41541-020-0194-5
- Harris VC, Armah G, Fuentes S, et al. Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. J Infect Dis. 2017;215(1):34–41. doi: 10.1093/infdis/jiw518
- Harris V, Ali A, Fuentes S, et al. Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes. 2018;9(2):93–101. doi: 10.1080/19490976.2017.1376162
- Parker EP, Praharaj I, Zekavati A, et al. Influence of the intestinal microbiota on the immunogenicity of oral rotavirus vaccine given to infants in south India. Vaccine. 2018;36(2):264–272. doi: 10.1016/j.vaccine.2017.11.031
- Fix J, Chandrashekhar K, Perez J, et al. Association between gut microbiome composition and rotavirus vaccine response among Nicaraguan infants. Am J Trop Med Hyg. 2020;102(1):213. doi: 10.4269/ajtmh.19-0355
- Huda MN, Lewis Z, Kalanetra KM, et al. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134(2):e362–e372. doi: 10.1542/peds.2013-3937
- de Koff EM, van Baarle D, van Houten MA, et al. Mode of delivery modulates the intestinal microbiota and impacts the response to vaccination. Nat Commun. 2022 Nov 15;13(1):6638. doi: 10.1038/s41467-022-34155-2
- Isolauri E, Joensuu J, Suomalainen H, et al. Improved immunogenicity of oral D x RRV reassortant rotavirus vaccine by lactobacillus casei GG. Vaccine. 1995;13(3):310–312. doi: 10.1016/0264-410X(95)93319-5
- Kukkonen K, Nieminen T, Poussa T, et al. Effect of probiotics on vaccine antibody responses in infancy–a randomized placebo-controlled double-blind trial. Pediatr Allergy Immunol. 2006 Sep;17(6):416–421.
- Soh SE, Ong DQR, Gerez I, et al. Effect of probiotic supplementation in the first 6 months of life on specific antibody responses to infant Hepatitis B vaccination. Vaccine. 2010 Mar 19;28(14):2577–2579. doi: 10.1016/j.vaccine.2010.01.020
- Youngster I, Kozer E, Lazarovitch Z, et al. Probiotics and the immunological response to infant vaccinations: a prospective, placebo controlled pilot study. Arch dischildhood. 2011;96(4):345–349. doi: 10.1136/adc.2010.197459
- Wu BB, Yang Y, Xu X, et al. Effects of Bifidobacterium supplementation on intestinal microbiota composition and the immune response in healthy infants. World J Pediatr. 2016 May;12(2):177–182.
- Grassly NC, Praharaj I, Babji S, et al. The effect of azithromycin on the immunogenicity of oral poliovirus vaccine: a double-blind randomised placebo-controlled trial in seronegative Indian infants. Lancet Infect Dis. 2016;16(8):905–914. doi: 10.1016/S1473-3099(16)30023-8
- Tsang AKL, Lee HH, Yiu SM, et al. Failure of phylogeny inferred from multilocus sequence typing to represent bacterial phylogeny. Sci Rep. 2017 Jul 3;7(1):4536. doi: 10.1038/s41598-017-04707-4
- Deleo FR, Chen L, Porcella SF, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci USA. 2014 Apr 1;111(13):4988–4993. doi: 10.1073/pnas.1321364111

