ABSTRACT
The Drosophila transgenic technology and fluorescent protein fusions are powerful tools to analyze protein expression patterns, subcellular localization and protein dynamics. Recently, the Drosophila transgenic technology has been improved by the highly efficient phiC31 site-specific integration system. Many new and improved fluorescent proteins with desirable advantages have been developed. However, the phiC31 system and the newly developed fluorescent proteins have not been systematically applied in Drosophila transgenic vectors. Here, we have constructed a modular toolset of C-terminal fluorescent protein fusion vectors based on phiC31 site-specific integration system for the generation of transgenic Drosophila lines. These cloning vectors contain a variety of fluorescent tags, including blue, cyan, green or red fluorescent proteins, photoactivatable or photoswitchable fluorescent proteins, fluorescent timers, photosensitizers and bimolecular fluorescence complementation tags. These vectors provide a range of transcriptional regulation options including UAST, UASP, UASC, LexAop, QUAS, Ubi, αTub67C and αTub84B promoters, and two screening marker options including white and vermilion gene. The vectors have been tested in vivo and can produce fluorescent chimeric proteins that are functional.
Introduction
The technique of germ line transformation is an invaluable tool for the study of gene expression, regulation and function in Drosophila melanogaster. More recently Drosophila transgenic technology has been further improved by using the site-specific PhiC31 integration system, which is more efficient than previous techniques [Citation1,Citation2]. Based on this method, the transgene insertions integrated at the same attP site are directly comparable and mapping is unnecessary.
The binary expression system is a versatile genetic tool in Drosophila. So far, several binary expression systems have been developed in Drosophila, including the Gal4/UAS, LexA/LexAop and QF/QUAS systems [Citation3–Citation5]. The Gal4/UAS system is the most widely used in Drosophila, consisting of two main components: the yeast Gal4 transcriptional activator and a transgene under the control of a UAS promoter. The pUAST and pUASP vectors are widely used for generating UAS transgenes, containing hsp70 basal promoter and P transposase promoter respectively. The pUAST vector allows for efficient expression in somatic cells, while the pUASP vector allows for maternal germline expression [Citation6]. Recently, the UASC vector has been developed, in which the hsp70 basal promoter was replaced by Drosophila Synthetic Core Promoter (DSCP) to avoid unwanted leaky expression [Citation7]. To enable parallel and controlled manipulation in multiple tissues and cell types, two other binary expression systems have been developed in Drosophila, including the LexA/LexAop system and the QF/QUAS system. The LexA/LexAop system is based on the bacterial LexA DNA-binding protein that regulates expression of transgenes fused to a LexA operator (LexAop) promoter [Citation4]. Recently, a LexA-based enhancer trap collection has been generated [Citation8], which facilitates the application of the LexA/LexAop system in neuroendocrine and developmental biology investigations. The QF/QUAS system, based on the regulatory gene QF from Neurospora crassa, is the newest binary expression system in Drosophila. Conceptually identical to the binary Gal4/UAS system, the QF/QUAS system uses the QUAS promoter and the transcriptional activator QF [Citation5]. Currently, there are few drivers available for the QF/QUAS system, limiting its usefulness. But new methods that allow for conversion of the GAL4 drivers into lexA or QF drivers have been found, they include the Homology-Assisted CRISPR Knock-in (HACK) method and the Integrase Swappable In Vivo Targeting Element (InSITE) system [Citation9,Citation10]. The availability of the three binary expression systems allows biologists to simultaneously perform two or three manipulations of gene expression in vivo, which could powerfully enhance studies of development, metabolism, and neurobiology in Drosophila. Besides the popular binary expression systems, ubiquitous expression promoters are also widely used in Drosophila transgenic expression, such as the Ubiquitin-63E promoter (Ubi) and the α-tubulin 84B promoter (αTub84B), which drive ubiquitous expression in germline tissues, embryos, larvae, pupae and adult flies [Citation11,Citation12]. For special experimental designs that require a transgene to be expressed only in the female germline and loaded maternally into the early embryo, α-tubulin 67C promoter (αTub67C) can be used, since it drives transcription exclusively in the nurse cells [Citation13].
Fluorescent proteins (FPs) have ubiquitous applications in biological research. FPs are used for a wide variety of purposes including as markers of gene expression, protein localization dynamics, markers of cell morphology, markers of subcellular organelles, indicators of cellular activity, and markers of protein interactions. Recently, many new FPs with desirable properties have been developed. These FPs span a wide range of colours including cyan, green and red fluorescent proteins, such as mTurquoise2, mTFP1, mClover, mKO and mRuby2 [Citation14–Citation17]. The brightness, photostability and maturing time have been significantly improved. PAGFP is a canonical photoconvertible fluorophore [Citation18]. SPAGFP and C3PAGFP are two new enhanced PAGFPs with improved diffusional and folding properties [Citation19]. Dronpa is a reversible photoswitchable GFP-like FP and suitable for tracking fast diffusion or transport of signalling molecules in live cells [Citation20]. Dendra2 and mEosFP are two green-to-red photoactivatable FPs and suitable for protein tracking in live cells [Citation21,Citation22]. SuperNova and miniSOG are photosensitizers that can be used for light inactivation of target proteins or photoablation of specific cell populations [Citation23,Citation24]. Besides fluorescent labelling, miniSOG fusion can also be used as a label for electron microscopy. Fluorescent timers, Fast-FT and Slow-FT, can be used to understand how protein localization changes over time from a single image [Citation25]. Though these new FPs have a variety of advantages in multicolour labelling, live imaging and protein tracking, they have not been systematically applied to Drosophila transformation vectors.
The PhiC31 transformation method in Drosophila has been developed for more than a decade [Citation1,Citation2]. However, several expression vectors for in vivo assays lack attB sites and are incompatible with the highly efficient PhiC31 method. Furthermore, some applications in Drosophila study require the use of wider variety of fluorescent protein tags. The newly developed FPs have not been systematically applied in Drosophila transgene vectors. The generation of a transgene construct expressing a chimeric fluorescent fusion protein still entails multiple cloning steps. So far, white gene is the most widely used selectable marker in Drosophila transformation. However, it has been reported that the exact gene dosage of white is important in behavioural studies [Citation26]. To address these limitations and simplify the cloning steps, we have generated a series of insulated phiC31 transformation vectors which both allow specific or uniform fluorescent fusion protein expression and enable a choice of fluorescent proteins with different spectral qualities. The series of vectors provide a range of transcriptional regulation options, including the UAST, UASP, UASC, LexAop, QUAS, Ubi, αTub67C and αTub84B promoters, two selected marker options (white and vermillion), and the optimal flexibility during multicolour fluorescent labelling and live-cell imaging in vivo. These vectors are compatible with a wide range of experiments, and will expand the facility and usefulness of fluorescent tags for protein function studies in Drosophila.
Results and discussion
Generation of the two starting vectors
We planned to systematically generate Drosophila transformation vectors with an attB site and various FP tags. To do so, we first built the starting vectors pB2GW and pB2GV (). The pB2GW vector was derived from pattB, but have been modified to include two gypsy insulators flanking the multiple cloning site. The gypsy insulator has been reported to effectively inhibit both chromosome position effects and cis-regulatory element modification [Citation27,Citation28]. The pB2GW vector contains an attB fragment that allows integration into the genome at attP landing sites, a mini-white gene that allows selection of transformed animals, and a loxP site that facilitates elimination of transgene markers via Cre recombinase-mediated excision when used in combination with ZH-attP landing sites [Citation2]. It has been reported that the exact gene dosage of white is important in behavioural studies [Citation26]. To circumvent this problem and to minimize the vector size, we generated the pB2GV vector from pB2GW by replacing the screening marker gene mini-white with vermilion (). The pB2GW/V (pB2GW and pB2GV) starting vectors can be used for genomic fragment transformation.
Figure 1. Overview of the vectors. Above: Map of the starting vectors pB2GW and pB2GV (to scale). The pB2GW vector contains a mini-white gene, a phiC31 integrase compatible attB sequence, a loxP site and an ampicillin resistance (ampR) gene. In pB2GV vector, the screening marker mini-white gene is replaced by vermilion gene. The multiple cloning site (MCS) is flanked by gypsy insulator sequences (In). The starting vectors are suitable for cloning genomic fragments. Below: Schematic of the parental vectors and fluorescent protein (FP) tagging vectors (not to scale). pKW/V and pKW/V-FP are suitable for expression of a gene of interest under the regulation of native promoter and enhancer elements. pUTSW/V, pUTSW/V-FP, pUCSW/V, pUCSW/V-FP, pUPKW/V and pUPKW/V-FP are suitable for Gal4 regulated transgene production. pQUSW/V and pQUSW/V-FP are suitable for QF regulated transgene production. pLASW/V and pLASW/V-FP are suitable for LexA regulated transgene production. pUbSW/V, pUbSW/V-FP, pUbKW/V, pUbKW/V-FP, pT84SW and pT84SW-FP are suitable for ubiquitous expression in germline tissues, embryos, larvae, pupae and adult flies. pT67SW/V and pT67SW/V-FP are suitable for female germline and early embryo expressions. Note that for vermilion version vectors, the XhoI site in MCS cannot be used in vector linearizing because the vermilion sequence contain an XhoI site. K10 pA: K10 polyadenylation signal; SV40 pA: SV40 polyadenylation signal; UAS: Upstream Activation Sequence; hsp70: hsp70 basal promoter; Delta2-3: Delta2-3 transposase promoter; QUAS: QF binding site; LexAop: LexA-binding site; Ubi: Ubiquitin-63E promoter; αTub67C: α-tubulin 67C promoter; αTub84B: α-tubulin 84B promoter; FP: Fluorescent Protein.
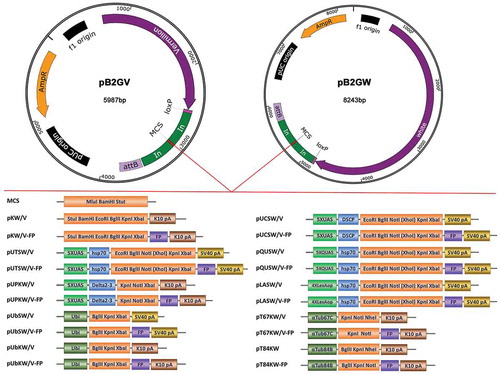
Generation of the 19 parental vectors
After generating the starting vectors, we introduced the K10 terminator into the starting vectors to generate the vectors pKW (pB2GW-K10) and pKV (pB2GV-K10), which allow expression of a gene of interest under the regulation of native promoter and enhancer elements (). K10 is a 3ʹUTR and terminator sequence from Drosophila gene fs(1)K10, which has previously been shown to support expression in both the maternal germline and zygotically [Citation6,Citation29]. To be compatible with all three binary expression systems (Gal4/UAS, LexA/LexAop and QF/QUAS), we generated pUTSW/V (pB2GW/V-UAS-hsp70-SV40), pUCSW/V (pB2GW/V-UAS-DSCP-SV40), pUPKW/V (pB2GW/V-UAS-Delta2-3-K10), pLASW/V (pB2GW/V-LexAop-hsp70-SV40), and pQUSW/V (pB2GW/V-QUAS-hsp70-SV40) vectors, which contains UAS-hsp70, UAS-DSCP, UAS-Delta2-3, LexAop-hsp70 and QUAS-hsp70 promoters respectively (). All vectors except for pB2GW/V-UASP contain the same multicloning site (EcoRI-BglII-NotI-XhoI-KpnI-XbaI), which allows for easy exchange of inserts among the vectors (). Using the three binary expression systems, we can simultaneously manipulate two or three gene expression in vivo. Besides the binary expression systems, we also generated ubiquitous expression vectors, including pUbSW/V (pB2GW/V-Ubi-SV40), pUbKW/V (pB2GW/V-Ubi-K10) and pT84KW (pB2GW-αTub84B-K10), which contain ubiquitin-63E promoter or α-Tubulin 84B promoter (). These promoters allow ubiquitous expression in germline tissues, embryos, larvae, pupae and adult flies [Citation11,Citation12]. For the female germline and the early embryo expression, we generated the pT67KW/V (pB2GW/V-αTub67C-K10) vectors, which contain the α-Tubulin 67C promoter. Overall, 19 parental vectors with different promoters and screening markers were generated ( and Table S1).
Generation of the C-terminal FP tagging vectors
After generating 19 parental vectors mentioned above, we cloned a number of FP tags into the parental vectors to generate a set of C-terminal FP tagging vectors (, and Table S1). These FP tags span a wide range of colours and desirable properties (), include one blue FP (EBFP2), 4 cyan FPs (mCerulean, mTurquoise, mTurquoise2, mTFP1), 3 green FPs (EGFP, mEGFP, mClover), 4 yellow FPs (Venus, mCitrine, mKO, mOrange), 5 red FPs (mCherry, TagRFP-T, tdTomato, mRuby, mRuby2), 6 photoactivatable or photoswitchable FPs (PAGFP, SPAGFP, C3PAGFP, Dronpa, mEosFP, Dendra2), 2 fluorescent timers (Slow-FT, Slow-FT), 2 photosensitizers (miniSOG, SuperNova) and 4 bimolecular fluorescence complementation (BiFC) tags based on EGFP or Venus (EGFP-N, EGFP-C, VNm9, VC155). The properties and references of these FPs are listed in . We also generated epitope tagged FPs (FP-HA, FP-Myc, FP-V5) for tagging of these vectors (Table S2). The epitope tagged FPs include HA, Myc or V5 tagged mCerulean, Venus, EGFP and mCherry (Table S2). FP-HA, FP-Myc and FP-V5 tags can be used for the detection of the tagged proteins by immunoblotting and immunofluorescence microscopy, and they can also be used to detect protein–protein interaction by co-immunoprecipitation. Photoactivatable GFP fusion proteins are not visible before photoactivation. We generated RFP-PAGFP tandem FP tags to make the tagging proteins trackable by red channel before photoactivation. The tandem FP tags include various RFPs (TagRFP-T, mCherry, tdTomato, mRuby, mRuby2) fused with different photoactivatable GFPs (PAGFP, SPAGFP, C3PAGFP) (Table S2). FlAsH-StrepII-TEV-3xFlag (FSTF) fused EGFP or PAGFP are multiple tags. The FlAsH in FSTF multi-tag can be recognized by specific di-arsenic compounds and become fluorescent within seconds upon binding [Citation30]. It is suitable for live cell labelling, protein affinity purification or electron-microscopic visualization. The StrepII-TEV-3xFlag in FSTF multi-tag can be used for tandem affinity purification which is useful for protein complexes analysis [Citation31]. In total, 546 vectors with various promoters, screening markers and FP tags were generated (Table S1).
Table 1. Properties and references of the fluorescent proteins.
Transgene examples using the vectors
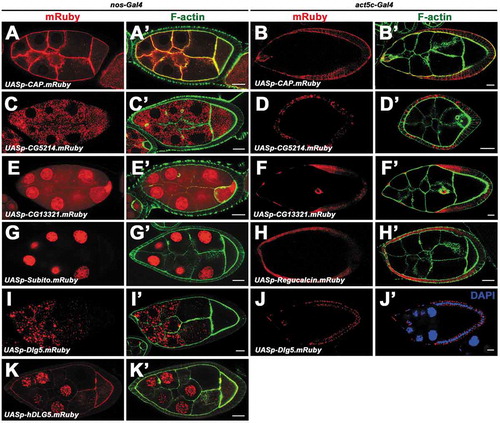
Examples of the utility of these vectors are illustrated with several transgenes listed in Table S4, including the transgenes of Discs large 5 (Dlg5), Cornetto (Corn), Cbl-associated protein (CAP), Receptor of activated protein kinase C 1 (Rack1), Regucalcin, Subito (Sub), CG5214, CG13321 and human DLG5 (hDLG5). The expression and localization of these proteins in Drosophila ovaries were examined ( and ). Dlg5 is an evolutionarily conserved membrane-associated guanylate kinase (MAGUK) family protein and is required for epithelial structure maintenance [Citation32]. We have previously shown Drosophila Dlg5 is required for apical polarity maintenance [Citation33]. The genomic construct Dlg5-TagRFP-T is based on pB2GW vector, containing 9.6kb genomic sequence that spans the whole dlg5 locus ()). The TagRFP-T fluorescent tag was inserted in-frame in the upstream of the stop codon. Dlg5-EGFP construct is based on the pKW-GFP vector, containing 1.7kb endogenous upstream regulatory sequence ()), Dlg5 CDS, EGFP tag and K10 terminator. Both of the constructs were transformed into the attP4 site and could rescue the lethality of dlg5 mutant (). They show a wide expression pattern of Dlg5 in Drosophila ovaries ()). Ubi-Dlg5.EGFP, Ubi-Dlg5.TagRFP-T and Ubi-Dlg5.mCherry transgenes can also rescue the lethality of dlg5 mutants (), revealing that the FP fused Dlg5 proteins are functional. Dlg5 proteins expressed by these constructs localizes in apical domains of follicle cells and nurse cell membranes in early stage egg chambers ()), which is consistent with the previous report [Citation33]. Ubi-Dlg5.EGFP shows ubiquitous expression of Dlg5 in egg chamber, eye disc, wing disc and embryo ()), indicating that the Ubi promoter is functional. UASt-Dlg5.mCerulean and UASt-Dlg5.mCherry are based on pUTSW-Cer and pUTSW-Ch vectors, respectively. Expression of UASt-Dlg5.mCerulean and UASt-Dlg5.mCherry as driven by act5C-Gal4 result in strong puncta aggregation in follicle cells of stage 9 egg chambers ()). This localization pattern is distinct from Ubi-Dlg5.EGFP which localizes in cell membranes ()), indicating that the accurate subcellular localization of a protein could be altered by overexpression, especially for proteins that have oligomerization potential. Dlg5-EGFP is useful in expression pattern analysis, while Ubi-Dlg5.EGFP is much brighter than Dlg5-EGFP and useful in subcellular localization analysis. The UASt-Dlg5.mCerulean and UASt-Dlg5.mCherry transgenes might be useful in gain-of-function analysis. CAP belongs to the Cbl-associated protein family and regulates junctional membrane and cytoskeletal organization [Citation34]. Ubi-CAP.EGFP and UASp-CAP.mRuby shows cytoskeleton-like localization in follicle cells and germline cells () and ’)). Corn is a microtubule-associated protein and binds to Myosin VI [Citation35]. UASt-Corn.mRuby and Ubi-Corn.EGFP shows vesicle-like accumulation in the apical region of follicle cells ()). This localization pattern is similar to the reported subcellular localization of Corn, which shows Corn protein is concentrated in the apical cytoplasm in epithelial cells of embryos [Citation36]. Rack1 is a scaffolding protein containing seven tandem WD1 motifs [Citation37]. UASt-Rack1.EGFP driven by slbo-Gal4 shows smeared localization in cytosol and significant enrichment in cell membrane ()). This localization pattern is similar to the reported Rack1 antibody staining which shows diffuse cytosolic localization and membrane localization [Citation38]. UASt-Dlg5.mEosFP and UASt-Dlg5.PAGFP.mRuby transgenes can be used in photoswitching experiment to convert the green protein puncta to red ones (’)), which will be useful in protein tracking. We also tested the pUTSW vector in S2 cells using UASt-Lifeact.mOrange and UASt-Lifeact.mClover constructs. Lifeact is a marker to label F-actin [Citation39]. The brightness and signal contrast of Lifeact.mClover are much stronger than those of Lifeact.mOrange. CG5214 is a predicted E2 member of the a-ketoglutarate dehydrogenase complex [Citation40]. UASp-CG5214.mRuby displays cytoplasm localization in puncta form (’)). CG13321 is a protein containing four DM9 repeats with unknown function. UASp-CG13321.mRuby shows nucleus and cytoplasm localization in germline cells and follicle cells (’)). Subito is a kinesin-like protein that is required for bundling microtubules [Citation41]. UASp-Subito.mRuby shows strong localization in nucleus (’)). The function of Regucalcin in Drosophila is unknown, but in mammals it functions in regulation of Ca2+ ion concentrations through modulation of the Ca2+ pumping activity [Citation42]. UASp-Regucalcin.mRuby shows diffused localization in cytoplasm and in cell nucleus (’)). UASp-Dlg5.mRuby expression results in strong protein aggregation in follicle cells and nurse cells (’)), which is distinct from the Ubi-Dlg5.EGFP localization and probably due to the overexpression by UAS/Gal4 system. UASp-hDLG5.mRuby shows cell membrane localization, ring canal localization and strong nucleus localization in germline cells (’)). Besides the transgene examples mentioned above, there are also published transgenes based on these vectors, such as Ubi-GFP and UASt-Sec3.EGFP [Citation43,Citation44]. Therefore, our vectors were effective in generating transgenic Drosophila lines for protein expression and localization analysis.
Table 2. Lethality rescue of dlg5 mutants by dlg5 transgenes.
Figure 2. Expression and localization of the transgene examples. a, Gene structure and mutant alleles of dlg5. Orange boxes represent the coding sequences, and grey boxes represent untranslated regions. The 1.7kb and 9.6kb genomic sequences for Dlg5-EGFP and Dlg5-TagRFP-T constructs are indicated respectively. b and c, Expression and localization of the full-length genomic construct Dlg5-TagRFP-T (b) and the mini genomic construct Dlg5-EGFP (c) in ovaries. d-g, Expression and localization of Ubi-Dlg5.EGFP in egg chamber (D), embryo (e), eye disc (f), and wing disc (g). h-k, Expression and localization of Ubi-Dlg5.TagRFP-T (h), Ubi-Dlg5.mCherry (i), Ubi-CAP.EGFP (j) and Ubi-Corn.EGFP (k) in different stage egg chambers. l-o, Expression and localization of UASt-Dlg5.mCerulean (l), UASt-Dlg5.mCherry (m), UASt-Rack1.EGFP (n, green) and UASt-Corn.Ruby (o) in stage 9 or stage 10 egg chambers. p-q’, Photoactivation of UASt-Dlg5.PAGFP.mRuby and UASt-Dlg5.mEosFP in border cells (p, p’) and follicle cells (q, q’) respectively. The circles (p, p’) or the boxes (q, q’) highlight the region before (Left) and after (Right) the UV laser irradiation in the same sample. ‘pre-active’ (p, q) and ‘post-active’ (p’, q’) are indicated. R and S, Expression and localization of UASt-Lifeact.mOrange and UASt-Lifeact.mClover in S2 Cells. All the UASt transgenes were driven by act5C-Gal4 except the UASt-Rack1.EGFP was driven by slbo-Gal4. Scale bars: 10 um in B-D, F, H-S; 50 um in E and G.
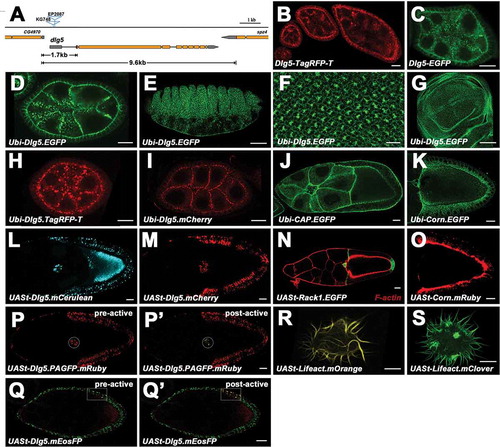
Figure 3. Expression and localization of the UASp transgenes in germline cells and follicle cells. a-k’, Expression and localization of UASp-CAP.mRuby (a-b’), UASp-CG5214.mRuby (c-d’), UASp-CG13321.mRuby (e-f’), UASp-Subito.mRuby (g, g’), UASp-Regucalcin.mRuby (h, h’), UASp-Dlg5.mRuby (i-j’), UASp-hDLG5.mRuby (k, k’) in germline cells (a, a’, c, c’, e, e’, g, g’, i, i’, k, k’) and follicle cells (b, b’, d, d’, f, f’, h, h’, j, j’). nos-Gal4 and act5C-Gal4 was used for germline expression and follicle cell expression respectively. Scale bars: 10um.
Different strategies for tagging Drosophila proteins have been described, such as the protein traps, the P[acman]-based recombineering-mediated genomic tagging, the Minos-Mediated Integration Cassette (MiMIC) technology and the homologous recombination (HR)-based gene targeting [Citation45–Citation49]. These approaches are excellent protein tagging tools, which tag proteins at their native genomic loci or in an endogenous genomic context. However, there are still many protein-coding genes in Drosophila which have not been tagged at their genomic loci. Utilizing the tagging vectors in this study, we could rapidly generate constructs in different expression levels, including overexpression using binary expression systems, moderate expression using ubiquitous promoters, and endogenous expression using endogenous promoters. Besides subcellular localization analysis, the constructs based on binary expression systems can also be used in overexpression and in combination with other genetic tools, such as the MARCM (mosaic analysis with a repressible cell marker) technique [Citation50]. Compared to the existing strategies, more FP options are provided in this study, but our strategy employ high-copy plasmids which are limited to inserts of large DNA fragments. We have successfully tested some vectors, such as pB2GW, pUTSW, pUPKW and pUbSW. Function of other vectors has to be test in specific studies although most of the genetic elements involved in these vectors have been utilized extensively in fly transgenesis.
In conclusion, we generated two starting vectors, 19 parental vectors with different promoters or screening markers, and 546 fluorescent protein tagging vectors with a wide range of fluorescent protein tagging. We expect that these reagents will expand the facility and usefulness of fluorescent tags for protein function studies in Drosophila, and will facilitate the proficiency and sophistication of Drosophila genetic analysis.
Materials and methods
The materials used and generated in this study will be available from the corresponding author JL upon request. For some vectors containing specific FPs, a copy of MTA (Material Transfer Agreement) between the user and the one who developed the FPs is needed.
Generation of the starting vectors pB2GW and pB2GV
To generate the pB2GW vector, the Gypsy insulator fragment was amplified from pCa4B2G [Citation51] with InsuS1 and InsuA1 primers, digested with BamHI/SpeI and ligated to the BamHI/NheI digested pattB vector [Citation2], resulting the pB1GW plasmid. The other Gypsy insulator fragment was amplified from pCa4B2G with InsuS2 and InsuA2 primers, digested with BamHI/SpeI and ligated to the BamHI/XbaI digested pB1GW plasmid, resulting the pB2GW vector. To generate the pB2GV vector, the vermilion marker gene was amplified from pValium20 [Citation52] using VerS and VerA primers. The other fragment was amplified from pB2GW using UBS and UBA primers to remove the white gene. The VerS/VerA primers has 18bp overlapping with the UBS/UBA primers at 5ʹ region. Then the two PCR products was ligated by recombination with the CloneEZ PCR cloning kit (Genescript), resulting the pB2GV vector. The sequences of the two starting vectors are available from the GenBank data libraries under accession no. MK424948 and MK424949. Primer sequences were listed in Table S5.
Generation of the 19 parental vectors
A K10 terminator fragment was amplified from pUASP with K10S and K10A primers, digested with MluI/BamHI and cloned into pB2GW digested similarly, resulting the pKW vector. The pUTSW vector was obtained through excision of the UAS-hsp70-MCS-SV40 sequence from plasmid pUAST with BamHI, and insertion of the resulting fragment into pB2GW. To generate the pUCSW vector, the synthetic core promoter, DSCP, was synthesized (Genescript), amplified with DSCPS and DSCPA primers and cloned to the pMD19-T simple vector (TAKARA), resulting the pMD19-DSCP construct. Then, the 5xUAS fragment was amplified from pUAST using UASS and UASA primers. The product was digested with HindIII/XbaI and cloned to the HindIII/NheI digested pMD19-DSCP, resulting the pMD19-UAS-DSCP plasmid. Finally, the UAS-DSCP fragment was released from the T vector and cloned to the pUTSW vector by EcoRI/HindIII digestion, resulting the pUCSW vector. The UAS-delta2-3-MCS-K10 fragment was amplified from pUASP with UPSL2 and K10A primers, then digested and cloned to the pUTSW vector by SbfI/MluI, resulting the pUPSW vector. To generate the pLASW vector, the hsp70 sequence was amplified from plasmid pUAST with hsS and hsA primers. The amplicon was then cloned into the pMD19-T simple T vector, resulting the pMD19-hsp70 construct. The LexAop fragment was amplified from pLOT [Citation4] using LexS and LexA primers, digested with HindIII/MluI and cloned into pMD19-hsp70. Finally, the LexAop-hsp70 fragment was amplified from the resulting plasmid and cloned into the HindIII/EcoRI digested pUTSW by recombination with the CloneEZ kit replacing the UAS-hsp70 sequence and resulting the pLASW vector. To generate the pQUSW vector, the QUAS-hsp70-MCS-SV40 fragment was amplified by two times primer extension PCR from pUAST using QS1/QA and QS2/QA primers. Then, the product was digested and cloned into pB2GW by BamHI, resulting the pQUSW vector. To generate the pUbSW vector, the Ubi promoter sequence was amplified from pUP2M plasmid [Citation11] and cloned into the SbfI/XhoI digested pUTSW by recombination with the CloneEZ kit replacing the UAS-hsp70 promoter and resulting the pUbSW vector. pUbKW was generated by digesting pUbSW with MluI/XbaI to remove the SV40 cassette and inserting the K10 fragment amplified from pUASP using K10S2 and K10A primers. To generate the pT67KW vector, the αTub67C promoter cassette was amplified from Canton-S flies using T67S and T67A primers and cloned into the SbfI/XbaI digested pUbKW by recombination with the CloneEZ kit replacing the Ubi promoter and resulting the pT67KW vector. Likewise, the αTub84B promoter containing vector pT84KW was generated using T84S and T84A primers. To generate the vermilion version of the pKW, pUTSW, pUCSW, pUPSW, pLASW, pQUSW, pUbSW, pUbKW and pT67KW vectors, the white gene was removed by PCR amplification from these vectors using UBS and UBA primers. The vermilion marker gene was amplified from pValium20 using VerS and VerA primers which has 18bp overlapping with the UBS/UBA primers at 5ʹ region. Then the two PCR products was ligated respectively by recombination with the CloneEZ PCR cloning kit, resulting the pKV, pUTSV, pUCSV, pUPSV, pLASV, pQUSV, pUbSV, pUbKV and pT67KV vectors. The sequences of the 19 parental vectors are available from the GenBank data libraries under accession no. MK424950 to MK424968 (Table S1). Primer sequences were listed in Table S5.
Generation of the C-terminal fluorescent protein tagging vectors
pUTSW, pUCSW, pLASW and pQUSW vectors were linearized by XhoI/XbaI. pKW, pUPSW, pUbSW, UbKW, pT67KW, pT84KW, pKV, pUTSV, pUCSV, pLASV, pQUSV, pUPSV, pUbSV, pUbKV and pT67KV vectors were linearized by KpnI/XbaI. Fluorescent proteins were amplified with specific primers, digested with XhoI/SpeI or KpnI/SpeI, and cloned to the linearized vectors, resulting the C-terminal fluorescent protein tagging vectors listed in the Table S1. The fluorescent proteins, specific PCR primers and templates were listed in Table S3. To generate PAGFP-mCherry, PAGFP-TagRFP-T or PAGFP-tdTomato tagging pUTSW vectors, PAGFP were amplified with GFPS8 and GFPA9 primers, digested with XhoI/SpeI and cloned into pUTSW-mCherry, pUTSW-TagRFP-T and pUTSW-tdTomato digested with XhoI/XbaI. To generate mCherry-C3PAGFP, TagRFP-T-C3PAGFP, tdTomato-C3PAGFP, mCherry-SPAGFP, TagRFP-T-SPAGFP and tdTomato-SPGFP tagging pUTSW vectors, mCherry and tdTomato were amplified with GFPS8 and GFPA9 primers. TagRFP-T were amplified with RTS2 and RTA2 primers. All the products were digested with XhoI/SpeI and cloned into pUTSW-C3PAGFP and pUTSW-SPAGFP digested with XhoI/XbaI. Primer sequences were listed in Table S5.
Expression constructs and transgenes
To generate C-terminal TagRFP-T tagged Dlg5 genomic construct, 1.4kb Dlg5 downstream sequence were amplified from the BAC CH321-38E02 (DGRC) with d3’S and d3ʹA primers, digested and cloned into the BamHI/MluI site of pB2GW, resulting pB2GW-d3ʹ. TagRFP-T was amplified and cloned into the NotI/XbaI site of pB2GW-d3ʹ, resulting pB2GW-TagRFP-T-d3ʹ. Then, 4.3kb dlg5 gene sequence were amplified with dG3’S/6509KA primers and cloned into the MluI/XbaI site of pB2GW-TagRFP-T-d3ʹ, resulting pB2GW-dlg5-TagRFP-T-d3ʹ. Finally, 3.9 kb fragment including 1.7 kb Dlg5 upstream sequence were amplified with 6509KS/d5ʹA2 primers, and cloned into the MluI/KpnI site of pB2GW-dlg5-TagRFP-T-d3ʹ, resulting the Dlg5-TagRFP-T genomic construct. To generate the Dlg5-EGFP construct, Dlg5 CDS were amplified from LD32687 (DGRC) with Dlg5S2/Dlg5A primers, cloned into BamHI/XbaI linearized pKW-GFP by the CloneEZ kit, resulting pKW-Dlg5CDS-GFP. Then, 1.7 kb dlg5 upstream sequence were amplified from CH321-38E02 with dBglS/d5ʹA4 primers, cloned into SbfI/XbaI linearized pKW-Dlg5CDS-GFP by the CloneEZ kit, resulting the Dlg5-EGFP construct. To generate the UASt-Dlg5.mCherry, UASt-Dlg5.mCerulean, UASt-Dlg5.PAGFP.mRuby, UASt-Dlg5.mEosFP, Ubi-Dlg5.EGFP, Ubi-Dlg5.mCherry, Ubi-Dlg5.TagRFP-T and UASp-Dlg5.mRuby constructs, Dlg5 CDS were amplified from LD32687 (DGRC), cloned into XhoI/XbaI linearized pUTSW-Ch, pUTSW-Cer, pUTSW-PAGRb, BglII linearized pUbSW-GFP, pUbSW-Ch, pUbSW-TRP, KpnI/XbaI linearized pUPKW-Rb, respectively, by the CloneEZ kit. Likewise, we generated the UASt-Corn.mRuby, UASt-Rack1.EGFP, UASt-Lifeact.mClover, UASt-Lifeact.mOrange, Ubi-Corn.EGFP, Ubi-CAP.EGFP, UASp-CG13321.mRuby, UASp-CG5214.mRuby, UASp-CAP.mRuby, UASp-Regucalcin.mRuby, UASp-Subito.mRuby and UASp-hDLG5.mRuby constructs using pUTSW-Rb, pUTSW-GFP, pUbSW-GFP, pUPKW-Rb vectors. The specific primers and CDS templates were listed in Table S4. All the constructs were sequenced and inserted into the attP2 docking site (Flybase ID FBti0040535) except the genomic Dlg5-TagRFP-T and Dlg5-EGFP constructs were inserted into the attP4 site (Flybase ID FBti0114366), the Ubi-Dlg5.TagRFP-T construct was inserted into the ZH-51C attP site (Flybase ID FBti0099697). All the transformations were performed using established PhiC31-based methods. UASt-Lifeact.mClover and UASt-Lifeact.mOrange constructs were used for S2 cell transfection and had not been transformed into the Drosophila genome.
Drosophila genetics
Flies were cultured following standard procedures at 25°C. The nos-Gal4 (Flybase ID FBti0015890), act5C-Gal4 (Flybase ID FBti0012293) and slbo-Gal4 (Flybase ID FBti0164789) tool lines and the dlg5KG748 mutant line (Flybase ID FBti0021658) were obtained from the Bloomington Drosophila Stock Center. The dlg5 mutant stock dlg5EP2087 (Flybase ID FBti0010837) was obtained from the Szeged stock Center.
Cell culture
Drosophila S2 cells were cultured in Schneider’s Drosophila medium (Gibco) supplemented with 10% foetal bovine serum (Gibco) and 1% penicillin/Streptomycin (Gibco) at 25°C. After 24 h culturing, PEI transfection method is used to transfect S2 cells [Citation53]. After 48 h, S2 cells were plated on concanavalin A (ConA)-coated coverslips and fixed in 7% formaldehyde according to standard protocols for confocal imaging.
Immunohisto chemistry and microscopy
Ovaries of adult female and imaginal discs of third instar larvae were dissected in phosphate-buffered saline (PBS) and then fixed in Devitellinizing buffer (7% formaldehyde) and heptane (Sigma) mixture (1:6) for 10 min. F-actin was labelled by Rhodamine phalloidin (1:100, Sigma). Photoactivation and photoconversion were performed manually by scanning of interested regions with 405 nm laser. The green and red fluorescent signals were acquired using 488 and 561 nm laser, respectively. Confocal images were obtained using a Leica TCS SP5 II or an Olympus FV1000 confocal microscope.
Supplemental Material
Download Zip (103.8 KB)Acknowledgments
We are grateful to Roger Y. Tsien, Norbert Perrimon, Konrad Basler, Johannes Bischof, Michele Markstein, Ulrich Nienhaus, Hiroki Oda, Soeren Diegelmann, Takeharu Nagai, Richard Binari, Richard Axel, and Drosophila Genomics Resource Center for reagents. We thank Bloomington Drosophila Stock Center and Szeged stock Center for fly stocks.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for the article can be accessed here.
Additional information
Funding
References
- Groth AC, Fish M, Nusse R, et al. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31 [Comparative study research support, U.S. Gov’t, P.H.S.]. Genetics. 2004 Apr;166(4):1775–1782. PubMed PMID: 15126397; PubMed Central PMCID: PMC1470814. eng.
- Bischof J, Maeda RK, Hediger M, et al. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases [Comparative study research support, Non-U.S. Gov’t]. Proc Natl Acad Sci U S A. 2007 Feb 27; 104(9):3312–3317. PubMed PMID: 17360644; PubMed Central PMCID: PMC1805588. eng
- Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002 Sep-Oct;34(1–2):1–15. PubMed PMID: 12324939.
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006 May;9(5):703–709. PubMed PMID: 16582903.
- Potter CJ, Tasic B, Russler EV, et al. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010 Apr 30;141(3):536–548. PubMed PMID: 20434990; PubMed Central PMCID: PMC2883883.
- Rorth P. Gal4 in the Drosophila female germline. Mech Dev. 1998 Nov;78(1–2):113–118. PubMed PMID: 9858703.
- Wang JW, Beck ES, McCabe BD. A modular toolset for recombination transgenesis and neurogenetic analysis of Drosophila. PubMed PMID: 22848718; PubMed Central PMCID: PMC3405054. eng PloS one. 2012;77:e42102.
- Pfeiffer BD, Ngo TT, Hibbard KL, et al. Refinement of tools for targeted gene expression in Drosophila [Research support, Non-U.S. Gov’t]. Genetics. 2010 Oct;186(2):735–755. PubMed PMID: 20697123; PubMed Central PMCID: PMC2942869. eng.
- Gohl DM, Silies MA, Gao XJ, et al. A versatile in vivo system for directed dissection of gene expression patterns. Nat Methods. 2011 Mar;8(3):231–237. PubMed PMID: 21473015; PubMed Central PMCID: PMC3079545.
- Lin CC, Potter CJ. Editing transgenic DNA components by inducible gene replacement in Drosophila melanogaster. Genetics. 2016 Aug;203(4):1613–1628. . PubMed PMID: 27334272; PubMed Central PMCID: PMC4981265.
- Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J Cell Sci. 2001 Feb;114(Pt 3):493–501. PubMed PMID: 11171319.
- O’Donnell KH, Chen CT, Wensink PC. Insulating DNA directs ubiquitous transcription of the Drosophila melanogaster alpha 1-tubulin gene. Mol Cell Biol. 1994 Sep;14(9):6398–6408. PubMed PMID: 8065369; PubMed Central PMCID: PMC359165.
- Matthews KA, Miller DF, Kaufman TC. Developmental distribution of RNA and protein products of the Drosophila alpha-tubulin gene family. Dev Biol. 1989 Mar;132(1):45–61. PubMed PMID: 2492961.
- Goedhart J, von Stetten D, Noirclerc-Savoye M, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun. 2012;3:751. PubMed PMID: 22434194; PubMed Central PMCID: PMC3316892.
- Ai HW, Henderson JN, Remington SJ, et al. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006 Dec 15;400(3):531–540. PubMed PMID: 16859491; PubMed Central PMCID: PMC1698604.
- Lam AJ, St-Pierre F, Gong Y, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012 Oct;9(10):1005–1012. PubMed PMID: 22961245; PubMed Central PMCID: PMC3461113.
- Karasawa S, Araki T, Nagai T, et al. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer [Comparative study research support, Non-U.S. Gov’t]. Biochem J. 2004 Jul 1;381(Pt 1):307–312. PubMed PMID: 15065984; PubMed Central PMCID: PMC1133789. eng.
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002 Sep 13;297(5588):1873–1877. PubMed PMID: 12228718; eng.
- Ruta V, Datta SR, Vasconcelos ML, et al. A dimorphic pheromone circuit in Drosophila from sensory input to descending output [Research Support, Non-U.S. Gov’t]. Nature. 2010 Dec 2;468(7324):686–690. PubMed PMID: 21124455; eng.
- Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004 Nov 19;306(5700):1370–1373. PubMed PMID: 15550670.
- Gurskaya NG, Verkhusha VV, Shcheglov AS, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol. 2006 Apr;24(4):461–465. PubMed PMID: 16550175.
- Wiedenmann J, Ivanchenko S, Oswald F, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion [Research Support, Non-U.S. Gov’t]. Proc Natl Acad Sci U S A. 2004 Nov 9;101(45):15905–15910. PubMed PMID: 15505211; PubMed Central PMCID: PMC528746. eng.
- Shu X, Lev-Ram V, Deerinck TJ, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011 Apr;9(4):e1001041. PubMed PMID: 21483721; PubMed Central PMCID: PMC3071375.
- Takemoto K, Matsuda T, Sakai N, et al. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation [Research Support, Non-U.S. Gov’t]. Sci Rep. 2013;3:2629. PubMed PMID: 24043132; PubMed Central PMCID: PMC3775092. eng.
- Subach FV, Subach OM, Gundorov IS, et al. Monomeric fluorescent timers that change color from blue to red report on cellular trafficking [Research Support, N.I.H., Extramural]. Nat Chem Biol. 2009 Feb;5(2):118–126. PubMed PMID: 19136976; PubMed Central PMCID: PMC2662996. eng.
- An X, Armstrong JD, Kaiser K, et al. The effects of ectopic white and transformer expression on Drosophila courtship behavior. J Neurogenet. 2000 Dec;14(4):227–43,271. PubMed PMID: 11342383.
- Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. Embo J. 1993 Feb;12(2):435–442. PubMed PMID: 8382607; PubMed Central PMCID: PMC413226.
- Kuhn EJ, Viering MM, Rhodes KM, et al. A test of insulator interactions in Drosophila. Embo J. 2003 May 15;22(10):2463–2471. PubMed PMID: 12743040; PubMed Central PMCID: PMC155999.
- Serano TL, Cheung HK, Frank LH, et al. P element transformation vectors for studying Drosophila melanogaster oogenesis and early embryogenesis. Gene. 1994 Jan 28;138(1–2):181–186. PubMed PMID: 8125300.
- Adams SR, Campbell RE, Gross LA, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002 May 29;124(21):6063–6076. PubMed PMID: 12022841.
- Kyriakakis P, Tipping M, Abed L, et al. Tandem affinity purification in Drosophila: the advantages of the GS-TAP system [Research Support, U.S. Gov’t, Non-P.H.S.]. Fly (Austin). 2008 Jul-Aug;2(4):229–235. PubMed PMID: 18719405; eng.
- Nechiporuk T, Fernandez TE, Vasioukhin V. Failure of epithelial tube maintenance causes hydrocephalus and renal cysts in Dlg5-/- mice. Dev Cell. 2007 Sep;13(3):338–350. PubMed PMID: 17765678; eng.
- Luo J, Wang H, Kang D, et al. Dlg5 maintains apical polarity by promoting membrane localization of Crumbs during Drosophila oogenesis. Sci Rep. 2016 May 23;6:26553. PubMed PMID: 27211898; PubMed Central PMCID: PMC4876392.
- Bharadwaj R, Roy M, Ohyama T, et al. Cbl-associated protein regulates assembly and function of two tension-sensing structures in Drosophila. Development. 2013 Feb 1;140(3):627–638. PubMed PMID: 23293294; PubMed Central PMCID: PMC3561792.
- Finan D, Hartman MA, Spudich JA. Proteomics approach to study the functions of Drosophila myosin VI through identification of multiple cargo-binding proteins. Proc Natl Acad Sci U S A. 2011 Apr 5;108(14):5566–5571. PubMed PMID: 21368190; PubMed Central PMCID: PMC3078346.
- Bulgheresi S, Kleiner E, Knoblich JA. Inscuteable-dependent apical localization of the microtubule-binding protein Cornetto suggests a role in asymmetric cell division [Research Support, Non-U.S. Gov’t]. J Cell Sci. 2001 Oct;114(Pt 20):3655–3662. PubMed PMID: 11707517; eng.
- McCahill A, Warwicker J, Bolger GB, et al. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002 Dec;62(6):1261–1273. PubMed PMID: 12435793.
- Kadrmas JL, Smith MA, Pronovost SM, et al. Characterization of RACK1 function in Drosophila development. Dev Dyn. 2007 Aug;236(8):2207–2215. PubMed PMID: 17584887.
- Riedl J, Crevenna AH, Kessenbrock K, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008 Jul;5(7):605–607. PubMed PMID: 18536722; PubMed Central PMCID: PMC2814344.
- Homem CCF, Steinmann V, Burkard TR, et al. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014 Aug 14;158(4):874–888. PubMed PMID: 25126791.
- Jang JK, Rahman T, Kober VS, et al. Misregulation of the kinesin-like protein Subito induces meiotic spindle formation in the absence of chromosomes and centrosomes. Genetics. 2007 Sep;177(1):267–280. PubMed PMID: 17660552; PubMed Central PMCID: PMC2013708.
- Fujita T, Inoue H, Kitamura T, et al. Senescence marker protein-30 (SMP30) rescues cell death by enhancing plasma membrane Ca(2+)-pumping activity in Hep G2 cells. Biochem Biophys Res Commun. 1998 Sep 18;250(2):374–380. PubMed PMID: 9753637.
- Zhu K, Liu M, Fu Z, et al. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet. 2017 Aug;13(8):e1006946. PubMed PMID: 28859085; PubMed Central PMCID: PMC5578494.
- Wan P, Wang D, Luo J, et al. Guidance receptor promotes the asymmetric distribution of exocyst and recycling endosome during collective cell migration. Development. 2013 Dec;140(23):4797–4806. PubMed PMID: 24198275; eng.
- Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond [Comparative study review]. Nat Rev Genet. 2001 Oct;2(10):756–768. PubMed PMID: 11584292; eng.
- Kelso RJ, Buszczak M, Quinones AT, et al. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D418–20. PubMed PMID: 14681446; PubMed Central PMCID: PMC308749.
- Venken KJ, Kasprowicz J, Kuenen S, et al. Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation [Research Support, Non-U.S. Gov’t]. Nucleic Acids Res. 2008 Oct;36(18):e114. PubMed PMID: 18676454; PubMed Central PMCID: PMC2566861. eng.
- Venken KJ, Schulze KL, Haelterman NA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes [Research support, N.I.H., Extramural research support, Non-U.S. Gov’t]. Nat Methods. 2011 Sep;8(9):737–743. PubMed PMID: 21985007; PubMed Central PMCID: PMC3191940. eng.
- Huang J, Zhou W, Dong W, et al. From the Cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering [Research support, N.I.H., Extramural research support, Non-U.S. Gov’t]. Proc Natl Acad Sci U S A. 2009 May 19; 106(20):8284–8289. PubMed PMID: 19429710; PubMed Central PMCID: PMC2688891. eng
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001 May;24(5):251–254. PubMed PMID: 11311363.
- Markstein M, Pitsouli C, Villalta C, et al. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.]. Nat Genet. 2008 Apr;40(4):476–483. PubMed PMID: 18311141; PubMed Central PMCID: PMC2330261. eng.
- Ni JQ, Zhou R, Czech B, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011 May;8(5):405–407. PubMed PMID: 21460824; PubMed Central PMCID: PMC3489273.
- Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7297–7301. PubMed PMID: 7638184; PubMed Central PMCID: PMC41326.
- Ai HW, Shaner NC, Cheng Z, et al. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007 May 22;46(20):5904–5910. PubMed PMID: 17444659.
- Rizzo MA, Piston DW. High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys J. 2005 Feb;88(2):L14–6. PubMed PMID: 15613634; PubMed Central PMCID: PMC1305173.
- Goedhart J, van Weeren L, Hink MA, et al. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods. 2010 Feb;7(2):137–139. PubMed PMID: 20081836.
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene. 1996;173(1 Spec No): 33–38. PubMed PMID: 8707053.
- Zacharias DA, Violin JD, Newton AC, et al. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.]. Science. 2002 May 3;296(5569):913–916. PubMed PMID: 11988576; eng.
- Nagai T, Ibata K, Park ES, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications [Research Support, Non-U.S. Gov’t]. Nat Biotechnol. 2002 Jan;20(1):87–90. PubMed PMID: 11753368; eng.
- Griesbeck O, Baird GS, Campbell RE, et al. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.]. J Biol Chem. 2001 Aug 3;276(31):29188–29194. PubMed PMID: 11387331; eng.
- Shaner NC, Campbell RE, Steinbach PA, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein [Comparative Study Evaluation Studies Letter Research Support, U.S. Gov’t, Non-P.H.S. Research Support, U.S. Gov’t, P.H.S.]. Nat Biotechnol. 2004 Dec;22(12):1567–1572. PubMed PMID: 15558047; eng.
- Shaner NC, Lin MZ, McKeown MR, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Nat Methods. 2008 Jun;5(6):545–551. PubMed PMID: 18454154; PubMed Central PMCID: PMC2853173. eng.
- Kredel S, Oswald F, Nienhaus K, et al. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PloS one. 2009;4(2):e4391. PubMed PMID: 19194514; PubMed Central PMCID: PMC2633614.
- Magliery TJ, Wilson CG, Pan W, et al. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005 Jan 12;127(1):146–157. PubMed PMID: 15631464.
- Saka Y, Hagemann AI, Piepenburg O, et al. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus [Research Support, Non-U.S. Gov’t]. Development. 2007 Dec;134(23):4209–4218. PubMed PMID: 17959720; PubMed Central PMCID: PMC2435607. eng.

