Abstract
The therapeutic effect of anti-cancer monoclonal antibodies stems from their capacity to opsonize targeted cancer cells with subsequent phagocytic removal, induction of antibody-dependent cell-mediated cytotoxicity (ADCC) or induction of complement-mediated cytotoxicity (CDC). The major immune effector cells involved in these processes are natural killer (NK) cells and granulocytes. The latter and most prevalent blood cell population contributes to phagocytosis, but is not effective in inducing ADCC. Here, we report that targeted delivery of the tumoricidal protein tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) to granulocyte marker C-type lectin-like molecule-1 (CLL1), using fusion protein CLL1:TRAIL, equips granulocytes with high levels of TRAIL. Upon CLL1-selective binding of this fusion protein, granulocytes acquire additional TRAIL-mediated cytotoxic activity that, importantly, potentiates antibody-mediated cytotoxicity of clinically used therapeutic antibodies (e.g., rituximab, cetuximab). Thus, CLL1:TRAIL could be used as an adjuvant to optimize the clinical potential of anticancer antibody therapy by augmenting tumoricidal activity of granulocytes.
Abbreviations
| ADCC | = | antibody-dependent cell-mediated cytotoxicity |
| ADCP | = | antibody-dependent cellular phagocytosis |
| CLL1 | = | C-type lectin-like molecule-1 |
| scFv | = | single-chain variable fragment |
| TRAIL | = | TNF-related apoptosis-inducing ligand |
Introduction
Therapeutic anti-cancer antibodies have become part of clinical practice and have significantly contributed toward improved patient survival in various malignancies.Citation1,2 Nevertheless, the efficacy of antibody-based anti-cancer approaches is still hampered by various mechanisms, including relapses of target antigen-negative disease, as well as intrinsic or acquired resistance to cytotoxic immune effector mechanisms.Citation3 Therefore, strategies that further augment the efficacy of antibody-based anti-cancer approaches are warranted.
The therapeutic effect of anti-cancer antibodies can derive from inhibition of target antigen-signaling and from their capacity to opsonize targeted cancer cells with subsequent phagocytic removal, induction of antibody-dependent cell-mediated cytotoxicity (ADCC) or induction of complement-mediated cytotoxicity (CDC). Immune effector cells involved in these processes are natural killer (NK) cells, monocytes/macrophages and granulocytes. Granulocytes, the most prevalent white blood cell, comprise ∼60% of all leukocytes in human blood. Granulocytes are professional phagocytes and efficiently remove target cells upon interaction of target cell-bound antibodies with their high affinity Fc receptors (FcR).Citation2,4 However, granulocytes are relatively weak inducers of (tumor) cell lysis via ADCC, especially when compared to NK cells.
Previously, we demonstrated that the tumoricidal activity of immune cells, specifically T cells, can be enhanced using the tumoricidal protein tumor necrosis factor-related apoptosis-inducing ligand (TRAIL).Citation5 TRAIL is expressed as type II transmembrane protein on NK-cells, where it is essential for NK cell–mediated immune surveillance of cancer cells.Citation6,7 Proteolytic processing of the extracellular domain of TRAIL generates a soluble form of TRAIL (sTRAIL) that retains prominent tumor-selective pro-apoptotic activity. Hence, recombinant sTRAIL is a promising effector molecule that is currently undergoing clinical evaluation.Citation8 By genetic fusion of sTRAIL to a T-cell selective antibody fragment, we previously showed that high levels of TRAIL could be delivered to the surface of T cells, hereby increasing their tumoricidal activity up to 500-fold.Citation5 In addition, a recent report demonstrated that coating leukocytes with TRAIL could trigger killing of cancer cells in the blood stream.Citation9 In line with this, we recently demonstrated that antibody fragment-mediated targeting of TRAIL to CD47 could improve the pro-phagocytic and apoptotic activity of leukocytes.Citation10
Here, we propose to augment the tumoricidal activity of granulocytes by surface delivery of high levels of TRAIL to C-type lectin-like molecule-1 (CLL1) using a CLL1-specific scFv antibody fragment. CLL1 is highly an selectively expressed on granulocytes, monocytes and dendritic cells, whereas other normal human cell types do not express CLL1.Citation11 Therefore, we hypothesized that CLL1:TRAIL would potentiate the tumoricidal activity of granulocytes and, moreover, would enhance the cytotoxic activity of therapeutic anticancer monoclonal antibodies.
Results
Construction of CLL1:TRAIL
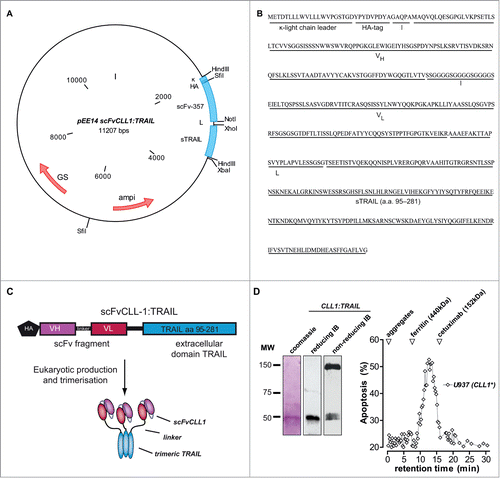
Construction of fusion protein CLL1:TRAIL was performed using the previously reported eukaryotic expression plasmid pEE14scFv:sTRAIL. This vector exploits the strong CMV promoter to drive recombinant protein expression and contains the murine kappa light-chain leader peptide, the hemagglutinin (HA) affinity tag and 2 multiple cloning sites (MCSs) separated by a 26 amino acid in-frame linker sequence (for vector map see ). In the first MCS, a 729 base pair DNA fragment encoding scFvCLL-1 (scFv-357) was directionally inserted using the unique SfiI and NotI restriction enzyme sites. The second MCS contained a PCR-truncated 593 base pair DNA fragment encoding the extracellular domain of human TRAIL (sTRAIL) (for sequence and features see ). The pEE14 vector encodes for the glutamine synthetase (GS) selection gene for protein expression in Chinese hamster ovary cells. This production platform produces and secretes correctly folded trimeric scFv:TRAIL fusion protein into the supernatantCitation12 and was here exploited for the production of CLL1:TRAIL at a concentration of 5.3 mg/L (for schematic representation of CLL1:TRAIL see ). Coomassie staining and immunoblot analysis of purified CLL1:TRAIL revealed a single band close to the predicted molecular weight of 51 kDa after reducing SDS-PAGE (), whereas in non-reducing conditions (non-reducing SDS-PAGE without sample boiling) CLL1:TRAIL predominantly migrated at a MW corresponding to that of trimeric CLL1:TRAIL (). Further, size exclusion FPLC of CLL1:TRAIL was performed, after which separate fractions were tested for apoptotic activity on CLL1-positive U937 cells (). This analysis confirmed that the apoptotic activity of CLL1:TRAIL is found in fractions that match a molecular weight closely corresponding to that of trimeric CLL1:TRAIL, with no evidence of CLL1:TRAIL protein aggregates ().
Figure 1. Construction of CLL1:TRAIL. (A) Vector map of scFvCLL1:TRAIL, designated CLL1:TRAIL. (B) Amino acid sequence of CLL1:TRAIL. (C) Schematic diagram of CLL1:TRAIL components and active CLL1:TRAIL trimer. (D) Gel stained with coomassie blue of reduced CLL1:TRAIL (β-mercaptoethanol and sample boiling) and corresponding immunoblot (IB) stained with anti-HA-HRP, as well as immunoblot analysis of CLL1:TRAIL (anti-HA-HRP) under nonreducing conditions without sample boiling. Size exclusion FPLC was performed, after which all collected fractions were tested for their potential to induce cytotoxicity in CLL1+ U937 cells. Apoptosis was determined by Annexin-V staining.
CLL1-resticted binding and apoptotic activity of CLL1:TRAIL
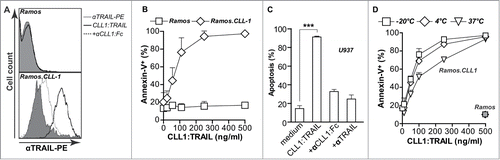
In earlier studies, it was shown that scFv:TRAIL fusion proteins bind via the scFv-domain to the target antigen of choice, whereupon membrane-associated TRAIL is capable of triggering apoptosis of target cells. CLL1:TRAIL was analogously designed to selectively bind to CLL1, leading to display of membrane-bound TRAIL on the surface, and subsequently to activation of TRAIL-receptor mediated cancer cell apoptosis. To verify this mode of action, we used a model system composed of parental CLL1-negative Ramos cells and CLL1 transfectant Ramos.CLL1 cells. Incubation of parental Ramos cells with CLL1:TRAIL (250 ng/ml) did not lead to detectable surface binding as evaluated by flow cytometric analysis with anti-TRAIL-PE antibody (, upper panel). However, incubation of Ramos.CLL1 cells with CLL1:TRAIL (250 ng/ml) at 0°C led to strongly increased fluorescent intensity upon TRAIL-PE staining, indicating high levels of surface-bound CLL1:TRAIL. Binding of CLL1:TRAIL was strongly inhibited by pre-incubation with epitope-competing CLL1 minibody (, lower panel, 2.5 μg/ml). In line with the lack of binding of CLL1:TRAIL to Ramos, treatment of these cells for 24 h with up to 500 ng/ml CLL1:TRAIL did not significantly induce apoptosis (, EC50 >500 ng/ml). In contrast, treatment of Ramos.CLL1 cells with CLL1:TRAIL potently induced apoptosis, with a maximum apoptotic effect of 97% at 500 ng/ml (, EC50 59.8 ng/ml). Further, on CLL1-positive U937 acute myeloid leukemia cells, CLL1:TRAIL triggered apoptosis that was abrogated by the presence of anti-CLL1 minibody or TRAIL-neutralizing antibody 2E5 (, EC50 36 ng/ml). Subsequently, CLL1:TRAIL was serum and temperature stable since it retained CLL1-restricted activity after storage at different temperatures (−20°C, 4°C, 37°C) in 5% FCS (). Thus, CLL1:TRAIL specifically binds to the CLL1 target antigen, whereupon the surface-associated TRAIL domain induced cancer cell apoptosis.
Figure 2. CLL1-resticted binding and apoptotic activity of CLL1:TRAIL. (A) Binding analysis of CLL1:TRAIL to Ramos (CLL1 negative) and Ramos.CLL1 (CLL1 positive) cells, with and without blocking with αCLL1:Fc using flow cytometry. (B) Target-restricted induction of apoptosis by increasing doses of CLL1:TRAIL was analyzed using Ramos and Ramos.CLL1. Apoptosis was analyzed using flow cytometric Annexin-V staining. (C) U937 (CLL1 positive) cells were treated with CLL1:TRAIL (250 ng/ml) in the presence or absence of αCLL1:Fc (2.5 μg/ml) or TRAIL neutralizing mAb2E5 (1 μg/ml) and apoptosis was determined using flow cytometric analysis of DioC6 staining. (D) CLL1:TRAIL was stored for up to 7 d in 5% FCS at −20°C, 4°C, and 37°C, after which CLL1-restricted activity was analyzed on the Ramos.CLL-1/Ramos cell line pair. Apoptosis was determined by Annexin-V staining. All graphs represent mean+SD. * p < 0.05, **p < 0.01, *** p < 0.001.
CLL1:TRAIL selectively binds to and augments tumoricidal activity of granulocytes
CLL1:TRAIL was produced with the aim of selectively equipping granulocytes with surface TRAIL to augment the cytotoxic potential of granulocytes. In line with literature, freshly isolated granulocytes expressed only minimal amounts of full-length trans-membrane TRAIL on the cell surface (). However, incubation with CLL1:TRAIL (250 ng/ml) markedly up-regulated TRAIL surface levels on granulocytes, an increase blocked by co-incubation with epitope-competing anti-CLL1 minibody (, 2.5 μg/ml). Further analysis revealed that CLL1:TRAIL enhanced the display of TRAIL on the surface of CD16+ granulocytes, but also to a lesser extent on the surface of CD14+ monocytes ().
Figure 3 (See previous page). CLL1:TRAIL selectively binds to and augments tumoricidal activity of granulocytes. (A) Flow cytometric analysis of CLL1:TRAIL binding to the surface of granulocytes, with and without αCLL1:Fc. (B) Within the granulocyte (CD16 positive) and monocyte (CD14 positive) population, the binding of CLL1:TRAIL to their cell surface was analyzed. (C) DLD-1 colon carcinoma cells (Target: T) were incubated with increasing amounts of leukocytes (Effector: E) (E:T ratio) in the presence or absence of CLL1:TRAIL (200 ng/ml) or MCSP:TRAIL (200 ng/ml) for 24 h, after which cell viability was assessed by MTS assay. (D) DLD-1 colon carcinoma cells were incubated with increasing concentrations of CLL1:TRAIL in the presence or absence of leukocytes (E:T = 5:1) and cell viability was assessed. CLL1 restricted activity of the fusion protein was evaluated by co-incubation with competing αCLL1:Fc minibody. (E) TRAIL- and caspase-dependent apoptosis was evaluated by co-incubating CLL1:TRAIL (200 ng/ml) and leukocytes (E:T = 5:1) with anti-TRAIL mab2E5 (1 μg/ml), and caspase-8 inhibitor zIETD-fmk (20 μM). (F) As in (C), but cytotoxicity was analyzed using LDH-release assay (G) DLD-1 colon carcinoma cells were incubated with CLL1:TRAIL (500 ng/ml) in co-cultures with increasing amounts of leukocytes or isolated granulocytes. Cell viability was determined using MTS assay. (H) Apoptosis induction of leukocytes (E:T = 10:1) and CLL1:TRAIL (200 ng/ml) was determined on DLD-1 3D-spheroids by Annexin-V staining on trypsin dissociated DLD-1 cells. All graphs represent mean+SD. * p < 0.05, **p < 0.01, *** p < 0.001, n.s. not significant.
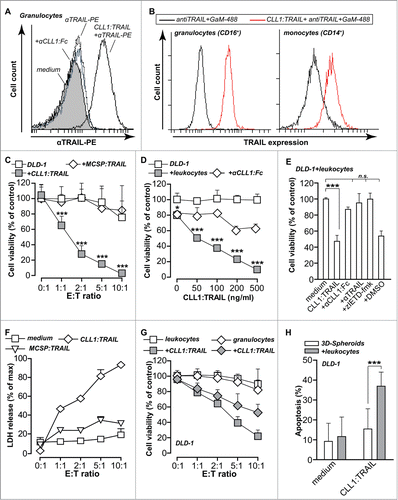
In line with the reported limited cytotoxic potential of granulocytes, mixing of DLD-1 cells with leukocytes only minimally affected DLD-1 viability after 24 h, with only 20% reduction in cell viability at the highest effector (E; leukocyte) to target (T; tumor cell) ratio evaluated (E:T-ratio=10:1, ). Treatment of CLL1-negative DLD-1 cells with CLL1:TRAIL (200 ng/ml) alone did not reduce cell viability (, see E:T ratio of 0:1), although at a higher concentration of 1 μg/ml CLL1:TRAIL did induce apoptosis (data not shown). However, treatment of mixed leukocyte/DLD-1 cultures with CLL1:TRAIL (200 ng/ml) strongly reduced the viability of DLD-1 cells in an E:T ratio dependent manner (). Control treatment with a TRAIL-fusion protein of irrelevant specificity (MCSP:TRAIL; 200 ng/ml) (), did not induce cell death, whereas MCSP:TRAIL did induce apoptosis in MCSP-positive melanoma cells (Supplementary Data S1A). Further, CLL1:TRAIL enhanced the cytotoxic potential of leukocytes (E:T-ratio=5:1) in a dose-dependent manner that was blocked by co-treatment with anti-CLL1 minibody (, 2 μg/ml). The tumoricidal effect of CLL1:TRAIL (200 ng/ml) in DLD-1/leukocyte co-cultures (E:T-ratio 5:1) was blocked by co-treatment with TRAIL-neutralizing antibody (mab2E5; 1 μg/ml), or caspase-8 inhibitor (zIETDfmk; 20 μM) (). Of note, the lactate dehydrogenase (LDH) release assay confirmed the results as obtained by MTS assays (). To confirm that granulocytes were the leukocyte population responsible for the enhanced cytotoxic effect by CLL1:TRAIL, the same experiments were performed with isolated granulocytes. Indeed, when CLL1:TRAIL (500 ng/ml) was added to mixed cultures of isolated granulocytes and DLD-1 cells, DLD-1 viability was strongly reduced (). Similar potentiating effects of CLL1:TRAIL (200 ng/ml) on granulocyte toxicity were observed when 3D-spheroid cultures of DLD-1 cells (E:T-ratio 10:1) were treated with this fusion protein ().
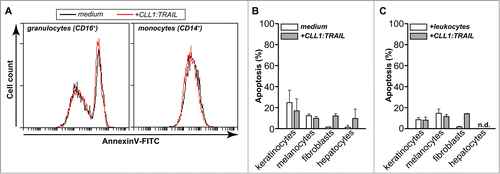
Importantly, CLL1:TRAIL (500 ng/ml) did not induce apoptosis in CLL-1 positive leukocyte populations, i.e., in granulocytes or monocytes (). Further, treatment of primary human keratinocytes, melanocytes, fibroblasts and hepatocytes with CLL1:TRAIL (700 ng/ml) alone (), or in mixed cultures with leukocytes () did not induce apoptosis. Thus, CLL1:TRAIL armed leukocytes, particularly granulocytes have enhanced tumoricidal activity without toxicity toward normal cells.
Figure 4. CLL1:TRAIL has no toxicity toward normal cells. (A) The effect of CLL1:TRAIL (500 ng/ml) on CLL-1 positive leukocytes was determined by assessing apoptosis (Annexin-V) by flow cytometry in CD16-positive granulocytes and CD14-positive monocytes, after 16 h treatment (using anti-CD14-APC, anti-CD16-PE-Dy647). Of note, leukocytes were stimulated with 10 ng/ml IFNγ to reduce spontaneous apoptosis which normally occurs in granulocytes. (B) Primary normal human cells were treated with CLL1:TRAIL (700 ng/ml) and apoptosis was measured by DioC6 assays. (C) As in G, in the presence of leukocytes (E:T = 10:1). n.d.: not determined
CLL1:TRAIL-armed leukocytes optimize the anti-cancer activity of therapeutic antibodies
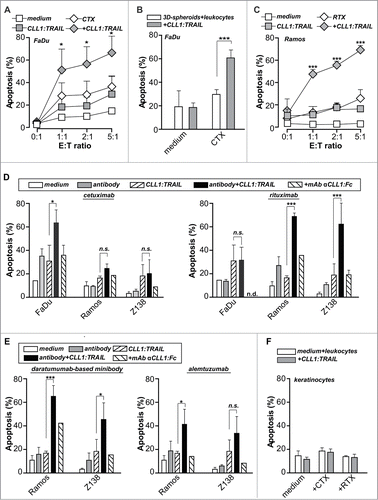
The enhanced tumoricidal activity of granulocytes armed with CLL1:TRAIL might also improve the ADCC-activity of clinically-approved antibodies. Indeed, co-treatment of EGFR-positive FaDu cells with EGFR-specific antibody cetuximab (CTX; 1 μg/ml) and CLL1:TRAIL-armed leukocytes (200 ng/ml) significantly enhanced pro-apoptotic activity compared to single-agent treatment (), also in established 3D-tumor cell spheres (). Similarly, co-treatment of Ramos B-cells with CLL1:TRAIL-armed leukocytes (200 ng/ml) and CD20-specific antibody rituximab (RTX; 1 μg/ml) significantly enhanced induction of apoptosis compared to leukocyte or rituximab treatment alone (). Control treatment of FaDu cells with rituximab and Ramos cells with cetuximab in combination with CLL1:TRAIL-armed leukocytes did not induce ADCC (). Further, CLL1:TRAIL-armed leukocytes (200 ng/ml) optimized the therapeutic activity of a humanized anti-CD38 antibody and anti-CD52 antibody alemtuzumab (1 μg/ml) on Ramos cells (). Similar results were obtained using B-cell lymphoma cell line Z138 for rituximab and anti-CD38 antibody (, E), but not for cetuximab and alemtuzumab as Z138 does not express EGFR or CD52. For all antibodies, the potentiating effect of CLL1:TRAIL-armed leukocytes was blocked by co-incubation with anti-CLL1 minibody (2,5 μg/ml) (). Importantly, combination treatment of primary human keratinocytes with CLL1:TRAIL-armed leukocytes and rituximab or cetuximab did not induce apoptosis (). Thus, binding of CLL1:TRAIL to CLL1-positive leukocytes potentiates anticancer ADCC activity of various clinically-approved antibodies without affecting normal healthy cells.
Figure 5 (See previous page). CLL1:TRAIL-armed leukocytes enhance the anti-cancer activity of therapeutic antibodies. (A) FaDu cells (EGFR positive) were incubated with different ratios of leukocytes and treated with cetuximab (CTX, 1 μg/ml), CLL1:TRAIL (200 ng/ml) combinations thereof. (B) FaDu cells cultured in 3D-spheroids were incubated with leukocytes (E:T = 10:1) and treated with cetuximab (CTX, 1 μg/ml), CLL1:TRAIL (200 ng/ml) or the combination. (C) Ramos cells (CD20 positive) were incubated with different ratios of leukocytes and treated with rituximab (RTX, 1 μg/ml), CLL1:TRAIL (200 ng/ml) or the combination. (D) As in A and C, using FaDu, Ramos and Z138 cells. In addition, αCLL1:Fc was used to block the effects of CLL1:TRAIL. (E) As in D, using Ramos and Z138 cells treated with the anti-CD38 antibody (daratumumab analog) and anti-CD52 antibody alemtuzumab (E:T ratio=5:1). (F) Primary normal human keratinocytes were treated with CLL1:TRAIL (700 ng/ml) and CTX or RTX (E:T ratio = 10:1). In all cases, apoptosis induction was determined using flow cytometric analysis of DioC6 staining. All graphs represent mean+SD. * p < 0.05, **p < 0.01, *** p < 0.001, n.s.: not significant, n.d.: not determined.
Discussion
In the current study, we report on fusion protein CLL-1:TRAIL that arms leukocytes, in particular granulocytes, with TRAIL. Hereby, CLL1:TRAIL enhanced the anti-tumor leukocyte activity per se and potentiated ADCC-activity of therapeutic anticancer antibodies. Thus, CLL1:TRAIL might be of clinical potential as adjuvant to optimize anticancer antibody-based therapy.
CLL1:TRAIL specifically augments the cytotoxic activity of granulocytes, a population of immune effector cells that has typically not attracted much attention for cancer therapy. However, granulocytes are an interesting target for immunotherapy, since they are the most abundant leukocyte population in the human blood (up to 60% of all leukocytes). Further, granulocytes have well-documented anticancer activity, with e.g., the administration of granulocytes reducing solid tumor growth and improving overall survival of tumor-bearing mice and rats.Citation13 Further, granulocytes are important anti-cancer immune effector cells in carcinoma, lymphoma and melanoma,Citation14,15 and are required for clinical activity of bladder cancer immunotherapy.Citation16
The most important function of granulocytes is that of phagocytosis of target cells. To a far lesser extent, granulocytes can contribute to ADCC. Both these functions are regulated by interactions between cell-surface FcR on granulocytes with the Fc of antibodies. Although granulocytes are very efficient phagocytes, their intrinsic potential to induce (tumor) cell lysis is only limited. Indeed, E:T ratios above 40:1 were needed to obtain significant monoclonal antibody-mediated lysis of cancer cells by granulocytes in previous studies, with only 20–35% cell death at an E:T ratio of 10:1.Citation17,18 In contrast, CLL1:TRAIL-armed granulocytes at the same E:T ratio triggered ∼90% apoptosis in DLD-1 cells without additional monoclonal antibodies, whereas >70% of FaDu cells were killed when CLL1:TRAIL-armed granulocytes were combined with CTX. Thus, CLL1:TRAIL-coated granulocytes can eliminate targeted cells not only by virtue of their intrinsic capacity for ADCC, but also by virtue of the cytotoxic activity conveyed by membrane-bound TRAIL. Of note, rational choice of the therapeutic antibody may contribute to the efficacy of TRAIL-mediated apoptosis by inhibition of target antigen-signaling. In this respect, we have previously published that an anti-EGFR:TRAIL fusion protein (termed scFv425:sTRAIL) can simultaneously inhibit EGFR-signaling and activate TRAIL-apoptotic signaling, yielding prominent tumoricidal activity both in vitro and in vivo.Citation19,20 Thus, combinatorial treatment with cetuximab and CLL1:TRAIL may be an attractive lead candidate strategy.
Enhancing the cytotoxic potential of granulocytes has also been attempted in previous studies. For instance, in vivo administration of human granulocyte-colony-stimulating factor (G-CSF) in patients upregulated the expression of high affinity receptor FcγRI on granulocytes, and enhanced their cytotoxicity toward lymphoma cells.Citation21 In addition, the administration of G-CSF strongly increased the amount of circulating granulocytes in lymphoma bearing mice, which resulted in enhanced ADCC when combined with administration of RTX.Citation22 These results have lead to the clinical evaluation of G-CSF and granulocyte-macrophage-colony-stimulating factor in the treatment of different cancer types.Citation23,24
Notably, the current study demonstrates that CLL1:TRAIL can significantly enhance the ADCC activity of granulocytes when combined with therapeutic antibodies, including rituximab, cetuximab, alemtuzumab and a daratumumab-based minibody. Therefore, CLL1:TRAIL might be included in existing treatment protocols using G-CSF and therapeutic anti-cancer antibodies, e.g., by co-injection of CLL1:TRAIL and antibodies, to optimize therapeutic efficacy. Alternatively, activated granulocytes could be isolated and ex vivo loaded with CLL1:TRAIL after multiple cycles of G-CSF and anti-cancer antibody and re-infused into the patient. Of note, ex vivo loading of T-cells with similar scFv:TRAIL fusion proteins strongly reduced in vivo tumor growth in mice.Citation5
In light of the in vivo use of CLL1:TRAIL it is important to note that the potentiating effect of CLL1:TRAIL on ADCC was target antigen specific, and will thus most likely not be associated with off-target and potentially toxic activity. Indeed, the application of non-binding antibodies did not enhance ADCC of TRAIL-loaded granulocytes. In addition, the combinational treatment of granulocytes, CLL1:TRAIL and different therapeutic antibodies was not cytotoxic toward primary normal human cells such as keratinocytes, melanocytes and fibroblasts. Further, TRAIL has proven to be safe in early clinical trials with no observations of dose-limiting toxicity.Citation25 Therefore, the use of CLL1:TRAIL in combination with therapeutic antibodies and possibly further combination with G-CSF may potentiate the anti-cancer efficacy of granulocytes in the absence of toxicity.
In conclusion, CLL1:TRAIL enhances leukocytic tumoricidal activity, particularly when combined with therapeutic anticancer antibodies like rituximab and cetuximab. Thus, CLL1:TRAIL might be of clinical potential as adjuvant to optimize anticancer antibody therapy in general.
Materials and Methods
Antibodies and reagents
Antibodies for flow cytometry used in this study were: PE-conjugated anti-TRAIL (Diaclone SAS, Besancon, France), anti-CD14-APC, anti-CD16-PE-Dy647, anti-CD16-FITC (Immunotools). Reagents used in this study where: TRAIL-neutralizing mAb 2E5 (Alexis, Kordia Life Sciences, Leiden, The Netherlands), recombinant human sTRAIL was from R&D systems (Oxon, UK), zIETDfmk was purchased from Calbiochem® (Merck Millipore). Cetuximab, rituximab, alemtuzumab were obtained from the hospital pharmacy. A CD38-targeted Daratumumab analog was generated in-house (see below).
Construction and production of CLL-1 minibody and Daratumumab analog
A DNA insert encoding scFvCLL-1 and scFvDaratumumab was synthesized based on published VH and VL DNA sequence data by DNA2.0 (Menlo Park, USA). Chimeric anti-CLL-1 and daratumumab minibodies were generated using the previously published anti-EpCAM minibody containing plasmid pEE14-MOC-Fc.Citation26 Briefly, the anti-EpCAM scFvMOC31 was swapped for scFvCLL-1 or scFvDaratumumab using unique SfiI and NotI restriction sites. Recombinant proteins were produced essentially as described before.Citation27
Cell lines and leukocytes
Leukemia cell lines U-937 (AML, CLL1+) and Ramos (B-CLL, CLL1−) were purchased from the ATCC (Manassas, USA). Carcinoma cell lines DLD-1 (colon), FaDu (hypopharyngeal), were also from ATCC. All cell lines were cultured in RPMI-1640 (Cambrex, New Jersey, New Hampshire, USA), supplemented with 10% (v/v) FCS. Primary human keratinocytes were cultured in serum free keratinocyte culture medium (BioWhittaker Europe SPRL, Belgium), human fibroblasts (NHDFjuv) were cultured in DMEM (Cambrex, New Jersey, New Hampshire, USA), supplemented with 10% (v/v) FCS, and primary human melanocytes were cultured in melanocyte growth medium (Cell Applications, Inc.) CLL1+ transfectant Ramos cells (Ramos.CLL1) were generated by electroporation with eukaryotic expression plasmid pCLL-1-IRIS2-EGFP. All above described cells were maintained at 37ºC/5% CO2.
After written informed consent, human leukocytes were obtained from venous blood from healthy volunteers using ammonium chloride lysis. Leukocytes were subsequently stimulated with IFN-γ (10 ng/ml, Immunotools) at a concentration of 1 × 106/ml for 1 h at 37°C. Isolated granulocytes were obtained by negative selection using MACS with anti-CD14/anti-CD3/anti-CD56/anti-CD19 microbead cocktail (Miltenyi Biotec).
Immunoblot analysis of CLL1:TRAIL
CLL1:TRAIL (1 μg) was separated by non-reducing SDS/PAGE (12% acrylamide) under reduing (β-mercaptoethanol and sample boiling) or nonreducing (without β-mercaptoethanol and sample boiling) and was subsequently electroblotted to nitrocellulose. Detection of CLL:TRAIL was performed by incubation with horseradish peroxidase-conjugated HA-tag antibody (Genscript), after which specific binding was visualized using chemoluminescence (WestDura Thermoscientific). Antibody incubation was performed at room temperature in PBS/5% skim milk for 1.5 h and followed by 3 washes with PBS/0,1%TWEEN. Coomassie blue staining of the gel was performed via standard protocol. In brief, the gel was stained with coomassie blue (Biorad) for 2 h, and subsequently destained (5% methanol, 7% Acetic acid in H2O).
Size-exclusion FPLC of CLL1:TRAIL
The solution behavior of CLL1:TRAIL was analyzed by size exclusion (SE) FPLC using a calibrated HiLoad 26/60 Superdex 200 Prep grade column (Amersham Biosciences AB, Upsala, Sweden). CLL1:TRAIL (5 ml) was loaded onto the column after which individual samples were collected at 1-minute intervals. All samples were analyzed for their capacity to induce apoptosis using the CLL1-positive and TRAIL sensitive cell line U937.
Flow cytometric analysis of cell surface TRAIL
For binding analysis of CLL1:TRAIL, CLL1 negative Ramos and Ramos.CLL1 cells were incubated with CLL1:TRAIL in the presence or absence of 10-fold molar excess of epitope-blocking CLL1 minibody, whereupon membrane expression of TRAIL was determined by incubating cells with anti-TRAIL-PE antibody at 4°C for 45 minutes. This was followed by 2 washes with excess PBS, after which binding was analyzed by flow cytometry on an Accuri C6 flow cytometer (BD biosciences). To analyze loading of TRAIL on the surface of specific leukocyte populations, leukocytes were incubated with CLL1:TRAIL and stained with anti-TRAIL-PE. In addition, anti-CD16 and anti-CD14 staining was performed to separately analyze TRAIL staining in granulocytes and monocytes, respectively.
Assessment of induction of cell death/loss of cell viability
Experiments were performed by mixing 3 × 104 tumor cells with leukocytes and/or fusion protein simultaneously in the E:T ratio/ concentrations as indicated. Of note, the E:T ratio of granulocytes and tumor cells was determined by flow cytometry based on FCS/SCC. Ratios were calculated by, after mixing leukocytes with tumor cell, gating on the granulocyte (Effector) and tumor cell (Target) populations (Fig. S1B). Thus, although all different leukocyte populations were present, the ratio was based on the number of granulocytes. Where indicated, competing CLL-1 minibody, anti-TRAIL mAb 2E5 or caspase-8 inhibitor zIETD-fmk or the various therapeutic antibodies were added to the mixed culture 30 min. prior to treatment with CLL1:TRAIL. After 24 h treatment, cell death was assessed by determining phosphatidylserine (PS) exposure on the outer membrane. PS on the outer membrane was analyzed by flow cytometry using AnnexinV-FITC (Immunotools, Friesoythe, Germany). In brief, all cells were harvested and washed with cold calcium binding buffer and resuspended in calcium buffer containing Annexin-V-FITC. After 10 min incubation at 4°C, PS-exposure was analyzed by flow cytometry using a BD Accuri C6 flow cytometer (BD Biosciences) and accessory CFlow Plus analysis software. Of note, tumor cells were gated based on FCS/ SCC, and AnnexinV-FITC binding was determined within this population. Correct separation was confirmed by fluorescent labeling of tumor cells, which enabled exclusion of granulocytes based on fluorescence (Fig. S1C) and, additionally, by labeling of granulocytes with CD16 antibody (Fig. S2). In brief, tumor cells were DiD labeled (Vybrant® DiD cell-labeling solution, Life technologies) by incubating 1 × 106 cells/ ml in serum free media with 5 μM DiD for 10 minutes at 37°C. Cells were washed to remove excess dye.
Loss of mitochondrial membrane potential (Δψ) was analyzed by flow cytometry using the cell-permeant green-fluorescent lipophilic dye DiOC6 (Molecular Probes, Eugene, USA). In brief, all cells were harvested and incubated for 20 min with DiOC6 (0.1 μM in medium) at 37°C. After washing, cells were resuspended in PBS and assessed by flow cytometry. Similar gating strategies as described for the determination of PS-exposure were used. Cell viability was measured using MTS according manufacturers protocol (CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay, Promega). In brief, 20% (v/v) MTS+PMS was added to culture medium of treated cells, and absorbance was measured at 490 nM after 1–2 hours of incubation at 37°C. Cells treated with 70% ethanol were included as a background control whereby the obtained absorbance was subtracted from all experimental conditions. Of note, tumor cells were carefully washed (3×) with PBS to remove the leukocytes before performing the MTS assay.
In addition, cell viability was determined using LDH-release assay (CytoTox 96® Non-Radioactive Cytotoxicity Assay, Promega) following manufacturers protocol. In brief, supernatants were collected, centrifuged to remove the death tumor cells and leukocytes, after which 50 μl was tranfered to a 96-wells plate. Subsequently, 50 μl of substrate mix was added to each sample and incubated for 30 min at room temperature. The reaction was stopped using 50 μl stop solution, and absorbance was measured at 490 nM. Of note, for each experiment appropriate controls were included (maximum release control, spontaneous release control, effector cell release conrol).
Formation and treatment of 3D-tumor spheres
3D-tumor spheres were generated by culture of 3·106 cells in low adherent culture flasks (Costar) for 72 hours. Subsequently, spheres were collected and separated from the medium by gravity, and the spheres were then used in similar experiments as described for single cell experiments. To assess the induction of cell death in the 3D-tumor spheres, spheres were harvested and washed with PBS. Subsequently, cells were dissociated using 10× trypsin-EDTA (Gibco) and then stained with DioC6 as described for single cell experiments.
Statistical analysis
Values are mean ± standard deviation (SD). Data was analyzed by Student's t-test, one or 2 way ANOVA+Tukey Kramer/Bonferroni post-test, respectively.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
suppl_figs_KMAB_1007811.zip
Download Zip (969.3 KB)Funding
This work was supported by Dutch Cancer Society grants RUG2009–4355 (EB), RUG2009–4542, RUG2011–5206, RUG2012–5541, RUG2013–6209 (to EB/WH), and the Netherlands Organization for Scientific Research (EB).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Adams GP, Weiner LM. Monoclonal antibody therapy of cancer. Nat Biotechnol 2005; 23:1147-57; PMID:16151408; http://dx.doi.org/10.1038/nbt1137
- Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer 2012; 12:278-87; PMID:22437872; http://dx.doi.org/10.1038/nrc3236
- Chouaib S, Meslin F, Thiery J, Mami-Chouaib F. Tumor resistance to specific lysis: a major hurdle for successful immunotherapy of cancer. Clin Immunol 2009; 130:34-40; PMID:19013109; http://dx.doi.org/10.1016/j.clim.2008.08.020
- Shuptrine CW, Surana R, Weiner LM. Monoclonal antibodies for the treatment of cancer. Semin Cancer Biol 2012; 22:3-13; PMID:22245472; http://dx.doi.org/10.1016/j.semcancer.2011.12.009
- de Bruyn M, Wei Y, Wiersma VR, Samplonius DF, Klip HG, van der Zee AG, Yang B, Helfrich W, Bremer E. Cell surface delivery of TRAIL strongly augments the tumoricidal activity of T cells. Clin Cancer Res 2011; 17:5626-37; PMID:21753155; http://dx.doi.org/10.1158/1078-0432.CCR-11-0303
- Cretney E, Takeda K, Yagita H, Glaccum M, Peschon JJ, Smyth MJ. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J Immunol 2002; 168:1356-61; PMID:11801676; http://dx.doi.org/10.4049/jimmunol.168.3.1356
- Takeda K, Hayakawa Y, Smyth MJ, Kayagaki N, Yamaguchi N, Kakuta S, Iwakura Y, Yagita H, Okumura K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat Med 2001; 7:94-100; PMID:11135622; http://dx.doi.org/10.1038/83416
- Fox NL, Humphreys R, Luster TA, Klein J, Gallant G. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) Receptor-1 and receptor-2 agonists for cancer therapy. Expert Opin Biol Ther 2010; 10:1-18; PMID:19857186; http://dx.doi.org/10.1517/14712590903319656
- Mitchell MJ, Wayne E, Rana K, Schaffer CB, King MR. TRAIL-coated leukocytes that kill cancer cells in the circulation. Proc Natl Acad Sci U S A 2014; 111:930-5; PMID:24395803; http://dx.doi.org/10.1073/pnas.1316312111
- Wiersma VR, He Y, Samplonius DF, van Ginkel RJ, Gerssen J, Eggleton P, Zhou J, Bremer E, Helfrich W. A CD47-blocking TRAIL fusion protein with dual pro-phagocytic and pro-apoptotic anticancer activity. Br J Haematol 2014; 164:304-7; PMID:24164421; http://dx.doi.org/10.1111/bjh.12617
- Marshall A, Willment J, Lin H, Williams DL, Gordon S, Brown GD. Identification and characterization of a novel human myeloid inhibitory C-type lectin-like receptor (MICL) that is predominantly expressed on granulocytes and monocytes. J Biol Chem 2004; 279:14792-802; PMID:14739280; http://dx.doi.org/10.1074/jbc.M313127200
- Bremer E, Kuijlen J, Samplonius D, Walczak H, de Leij L, Helfrich W. Target cell-restricted and -enhanced apoptosis induction by a scFv:sTRAIL fusion protein with specificity for the pancarcinoma-associated antigen EGP2. Int J Cancer 2004; 109:281-90; PMID:14750182; http://dx.doi.org/10.1002/ijc.11702
- Jaganjac M, Poljak-Blazi M, Kirac I, Borovic S, Joerg Schaur R, Zarkovic N. Granulocytes as effective anticancer agent in experimental solid tumor models. Immunobiology 2010; 215:1015-20; PMID:20122752; http://dx.doi.org/10.1016/j.imbio.2010.01.002
- Souto JC, Vila L, Bru A. Polymorphonuclear neutrophils and cancer: intense and sustained neutrophilia as a treatment against solid tumors. Med Res Rev 2011; 31:311-63; PMID:19967776; http://dx.doi.org/10.1002/med.20185
- Challacombe JM, Suhrbier A, Parsons PG, Jones B, Hampson P, Kavanagh D, Rainger GE, Morris M, Lord JM, Le TT, et al. Neutrophils are a key component of the antitumor efficacy of topical chemotherapy with ingenol-3-angelate. J Immunol 2006; 177:8123-32; PMID:17114487; http://dx.doi.org/10.4049/jimmunol.177.11.8123
- Brincks EL, Risk MC, Griffith TS. PMN and anti-tumor immunity-the case of bladder cancer immunotherapy. Semin Cancer Biol 2013; 23:183-9; PMID:23410637; http://dx.doi.org/10.1016/j.semcancer.2013.02.002
- Stockmeyer B, Beyer T, Neuhuber W, Repp R, Kalden JR, Valerius T, Herrmann M. Polymorphonuclear granulocytes induce antibody-dependent apoptosis in human breast cancer cells. J Immunol 2003; 171:5124-9; PMID:14607911; http://dx.doi.org/10.4049/jimmunol.171.10.5124
- Horner H, Frank C, Dechant C, Repp R, Glennie M, Herrmann M, Stockmeyer B. Intimate cell conjugate formation and exchange of membrane lipids precede apoptosis induction in target cells during antibody-dependent, granulocyte-mediated cytotoxicity. J Immunol 2007; 179:337-45; PMID:17579054; http://dx.doi.org/10.4049/jimmunol.179.1.337
- Bremer E, Samplonius DF, van GL, Dijkstra MH, Kroesen BJ, de Leij LF, Helfrich W. Simultaneous inhibition of epidermal growth factor receptor (EGFR) signaling and enhanced activation of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor-mediated apoptosis induction by an scFv:sTRAIL fusion protein with specificity for human EGFR. J Biol Chem 2005; 280:10025-33; PMID:15644326; http://dx.doi.org/10.1074/jbc.M413673200
- Bremer E, van Dam G, de Bruyn M, van Riezen M, Dijkstra M, Kamps G, Helfrich W, Haisma H. Potent systemic anticancer activity of adenovirally expressed EGFR-selective TRAIL fusion protein. Mol Ther 2008; 16:1919-26; PMID:18813279; http://dx.doi.org/10.1038/mt.2008.203
- Repp R, Valerius T, Sendler A, Gramatzki M, Iro H, Kalden JR, Platzer E. Neutrophils express the high affinity receptor for IgG (Fc gamma RI, CD64) after in vivo application of recombinant human granulocyte colony-stimulating factor. Blood 1991; 78:885-9; PMID:1714327
- Hernandez-Ilizaliturri FJ, Jupudy V, Reising S, Repasky EA, Czuczman MS. Concurrent administration of granulocyte colony-stimulating factor or granulocyte-monocyte colony-stimulating factor enhances the biological activity of rituximab in a severe combined immunodeficiency mouse lymphoma model. Leuk Lymphoma 2005; 46:1775-84; PMID:16263581; http://dx.doi.org/10.1080/17402520500182329
- Cheung IY, Hsu K, Cheung NK. Activation of peripheral-blood granulocytes is strongly correlated with patient outcome after immunotherapy with anti-GD2 monoclonal antibody and granulocyte-macrophage colony-stimulating factor. J Clin Oncol 2012; 30:426-32; PMID:22203761; http://dx.doi.org/10.1200/JCO.2011.37.6236
- Roche MR, Rudd PJ, Krasner CN, Matulonis UA, Berlin ST, Lee H, Silver M, Tran CD, Seiden MV, Penson RT. Phase II trial of GM-CSF in women with asymptomatic recurrent mullerian tumors. Gynecol Oncol 2010; 116:168-72; PMID:19922985; http://dx.doi.org/10.1016/j.ygyno.2009.10.075
- Herbst RS, Eckhardt SG, Kurzrock R, Ebbinghaus S, O'Dwyer PJ, Gordon MS, Novotny W, Goldwasser MA, Tohnya TM, Lum BL, et al. Phase I dose-escalation study of recombinant human Apo2L/TRAIL, a dual proapoptotic receptor agonist, in patients with advanced cancer. J Clin Oncol 2010; 28:2839-46; PMID:20458040; http://dx.doi.org/10.1200/JCO.2009.25.1991
- Helfrich W, Haisma HJ, Magdolen V, Luther T, Bom VJ, Westra J, van der Hoeven R, Kroesen BJ, Molema G, de Leij L. A rapid and versatile method for harnessing scFv antibody fragments with various biological effector functions. J Immunol Methods 2000; 237:131-45; PMID:10725458; http://dx.doi.org/10.1016/S0022-1759(99)00220-3
- de Bruyn M, Rybczynska AA, Wei Y, Schwenkert M, Fey GH, Dierckx RA, van Waarde A, Helfrich W, Bremer E. Melanoma-associated Chondroitin Sulfate Proteoglycan (MCSP)-targeted delivery of soluble TRAIL potently inhibits melanoma outgrowth in vitro and in vivo. Mol Cancer 2010; 9:301; PMID:21092273; http://dx.doi.org/10.1186/1476-4598-9-301

