ABSTRACT
Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6), a cell surface receptor, is expressed on normal epithelial tissue and highly expressed in cancers of high unmet medical need, such as non-small cell lung, pancreatic, and colorectal cancer. CEACAM receptors undergo homo- and heterophilic interactions thereby regulating normal tissue homeostasis and angiogenesis, and in cancer, tumor invasion and metastasis. CEACAM6 expression on malignant plasma cells inhibits antitumor activity of T cells, and we hypothesize a similar function on epithelial cancer cells.
The interactions between CEACAM6 and its suggested partner CEACAM1 on T cells were studied. A humanized CEACAM6-blocking antibody, BAY 1834942, was developed and characterized for its immunomodulating effects in co-culture experiments with T cells and solid cancer cells and in comparison to antibodies targeting the immune checkpoints programmed cell death protein 1 (PD-1), programmed death-ligand 1 (PD-L1), and T cell immunoglobulin mucin-3 (TIM-3).
The immunosuppressive activity of CEACAM6 was mediated by binding to CEACAM1 expressed by activated tumor-specific T cells. BAY 1834942 increased cytokine secretion by T cells and T cell-mediated killing of cancer cells. The in vitro efficacy of BAY 1834942 correlated with the degree of CEACAM6 expression on cancer cells, suggesting potential in guiding patient selection. BAY 1834942 was equally or more efficacious compared to blockade of PD-L1, and at least an additive efficacy was observed in combination with anti-PD-1 or anti-TIM-3 antibodies, suggesting an efficacy independent of the PD-1/PD-L1 axis.
In summary, CEACAM6 blockade by BAY 1834942 reactivates the antitumor response of T cells. This warrants clinical evaluation.
Introduction
Many cancers express immune checkpoint molecules (ICMs) to block T cell functions, limiting the efficacy of cancer immunotherapy.Citation1–3 However, T cell function can be reactivated through blockade of ICMs.Citation4 For example, several approved drugs targeting the programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) axis have achieved impressive clinical responses in several cancer types.Citation5 Still, most cancers do not respond to PD-1/PD-L1 inhibitors or they develop resistance to them.
Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6, CD66c, nonspecific cross-reacting antigen, NCA, or NCA-50/90) is a glycosylphosphatidylinositol (GPI)-linked cell surface protein that belongs to the CEACAM family of the immunoglobulin (Ig) supergene family.Citation6,Citation7 CEACAMs participate in the regulation of cell-cell adhesion, tissue formation, angiogenesis, apoptosis, tumor suppression, invasion, and metastasis formation but can also act as pathogen receptors.Citation8–10 They undergo homo- and heterophilic interactions with other CEACAMs, and CEACAM6 has been found to bind CEACAM5 and CEACAM8.Citation6,Citation7,Citation11 Since neither CEACAM5 nor CEACAM8 possesses intracellular signaling domains or is expressed on T cells, other CEACAM ligands may mediate the T cell suppression observed with CEACAM6. Among them, CEACAM1 is found on activated T cells and one of the few CEACAM receptors containing intracellular immunoreceptor tyrosine-based inhibitory motive (ITIM) domains for T cell signaling.Citation11 Indeed, CEACAM1-CEACAM1 homophilic interactions between T cells and CEACAM1-expressing cancer cells can play a role in antitumor T cell suppression, and binding of CEACAM1 by pathogens can lead to T cell inhibition.Citation9,Citation10
Human and mouse T lymphocytes in blood or lymph nodes generally express the main ITIM-containing CEACAM1-L isoforms CEACAM1–3 L and −4 L which are significantly upregulated at the surface in response to interleukin-2 (IL-2).Citation12 Apart from T cell inhibition, these isoforms may mediate adhesion of T cells to other lymphocytes or tumor cells, which contributes to their cytolytic function as well.Citation12 The isoform expressed most dominantly on T lymphocytes is CEACAM1-L, while CEACAM1–3S and −4S isoforms are expressed in much lower amounts.Citation13
Here, we investigated whether CEACAM6 represents an alternative ICM in immunotherapy-resistant human cancers which inhibit T cell responses independently of the PD-1/PD-L1 axis. CEACAM6 suppresses cytotoxic T cell responses against malignant human plasma cellsCitation14 and is expressed in cancer types largely resistant to PD-1/PD-L1 blockade, including colon and pancreatic cancer.Citation15–19 In these cancers, CEACAM6 levels correlate with tumor progression and adverse clinical outcomeCitation16,Citation17,Citation20–23 and CEACAM6 is more abundant than carcinoembryonic antigen (CEA, CEACAM5) in breast, pancreatic, mucinous ovarian, gastric and large adenocarcinoma, whereas in prostate cancer, CEACAM6 and CEA are expressed in equal amounts.Citation6,Citation15 Furthermore, CEACAM6 is expressed on granulocytes and macrophages, which constitute a major component of immunosuppressive tumor microenvironments.Citation24 Accordingly, CEACAM6 expression correlates with reduced T cell infiltration during colon cancer development.Citation25 In healthy conditions, CEACAM6 is expressed on myeloid cells in blood with the highest levels found on granulocytes,Citation26 resident myeloid cells,Citation24 and epithelial cells in the lungCitation27 and intestine,Citation15,Citation28 where it is often co-expressed with CEACAM1 and CEACAM5Citation15 and it contributes to normal tissue homeostasis with CEACAM7 for which an inverse correlation of expression with CEACAM6 has been found.Citation29 In cancer, this co-expression is deregulated, and indeed, CEACAM1, CEACAM5, and CEACAM6 are now considered valid clinical tumor biomarkers and promising therapeutic targets in melanoma, lung, colorectal, and pancreatic cancers.Citation6,Citation15,Citation29
CEACAM6 orthologs exist in human and non-human primates but not in rodents,Citation30,Citation31 which precludes the option to study CEACAM6 interactions in vivo in mouse or rat models.
Here, we show that the interaction between CEACAM6 on human solid cancer cells and CEACAM1 on activated tumor-reactive T cells suppresses the antitumor functions of T cells independently of the PD-1/PD-L1 axis. We further describe a humanized anti-CEACAM6 antibody BAY 1834942, which blocks this interaction and restores the antitumor activity of T cells in CEACAM6-positive cancers.
Methods
Antibody compounds and analysis tools
BAY 1834942 (anti-CEACAM6 antibody, human IgG2) was generated at Bayer AG (Germany) by immunizing mice with recombinant cynomolgus monkey CEACAM6 and subsequent humanization as described in detail in the Supplementary Methods.
The variable domains of the murine anti-CEACAM6 antibody 9A6 (Aldevron)Citation32,Citation33 were cloned from its hybridoma to generate a human IgG2 chimeric antibody. The anti-PD-L1 (human IgG2) and the anti-PD-1 (human IgG4 Pro) monoclonal antibodies (mAb) were generated at Bayer AG by cloning the variable domains of atezolizumab and nivolumab, respectively. The anti-CEACAM8 antibody (mouse IgM1 kappa, #555723) was purchased from BD Pharmingen. The anti-CEACAM5/6 antibody h16 C3 (as described in US20130189268,Citation34 human IgG1,Citation35,Citation36), the anti-human CEACAM1 antibody (as described in WO2013054320,Citation37 human IgG1 (TPP-3006) and human IgG2 (TPP-9145)) and isotype control antibodies TPP-1238 (huIgG2), TPP-1240 (huIgG4-S228P), and TPP-754 (huIgG1) were generated at Bayer AG. MAB2365 (an antibody against T cell immunoglobulin mucin-3 (TIM-3)) and its isotype control MAB006 are both recombinant rat IgG2a antibodies and were purchased from R&D Systems.
A list of additional tool antibodies used in the study can be found in Suppl. Table S1. In single experiments, antibodies were used as modified (chimeric) isotype versions, which is indicated accordingly.
Recombinant proteins
Recombinant human CEACAM1, −5, −6 proteins were purchased from R&D Systems, human CEACAM3 was obtained from Sino Biological. Other recombinant human or cynomolgus CEACAM6 variants including Fc fusions and domain variants were produced as described in the Supplementary Methods.
Cell lines and primary immune cells
The cell lines used in this study are summarized in Suppl. Table S2. The cell lines were obtained between 2002 and 2013 and authenticated using short tandem repeat DNA finger-printing at Leibniz Institute DSMZ-German Collection of Micro-organisms and Cell Cultures (DSMZ) before the experiments. For experimental studies, cell lines were harvested at 60–80% confluency. Detailed information about the cancer cell lines used and about their genetic modifications is provided in the Supplementary Methods.
The isolation of peripheral blood mononuclear cells (PBMCs) is described in the Supplementary Methods.
Tumor antigen-specific (i.e. survivin peptide-specific) T cells were generated from PBMCs of healthy donors as previously described.Citation38,Citation39 As survivin T cells do not express PD-1, PD1-expressing virus (influenza FluM1 peptide)-specific T cells were used as effector cells for testing the efficacy of the anti-PD-L1 and anti-PD-1 antibodies. Their generation is described in the Supplementary Methods.
Tumor-infiltrating lymphocytes (TILs) from pancreatic cancer (University Hospital Heidelberg Biobank, kindly provided by Dr. Isabel Poschke at the German Cancer Research Center, DKFZ, Heidelberg), myeloma TILs, and lung cancer TILs were prepared as described in the Supplementary Methods.
Jurkat E6.1 cells (ATCC) and Jurkat T-REx (Bayer AG) cells were used to generate CEACAM1 knockout T cells for experiments. The details of their generation are described in the Supplementary Methods.
Ethical statement
The studies on human samples were conducted according to the ethical principles of research with human participants (including the revised Helsinki Declaration). The study protocols were approved by the local ethics committees with written informed consent obtained from each volunteer.
CEACAM6 protein expression in normal and cancerous tissues and leukocytes
CEACAM6 protein expression was determined in tissue microarray (TMA) slides of various primary tumor and normal tissue samples with the anti-human CEACAM6 antibody 9A6 mIgG1 (Aldevron) and BAY 1834942 (or a mIgG2a variant thereof) by immunohistochemistry (IHC) as described in the Supplementary Methods. The paraffin-embedded samples were obtained from Provitro (Berlin, Germany), Asterand Bioscience (Royston, UK), and Indivumed (Hamburg, Germany). For comparison of CEACAM6 and PD-L1 expression in non-small cell lung cancer (NSCLC) tissue, samples were stained with an anti-PD-L1 antibody (1:150, Abcam, EPR1161Citation2) in a BOND automated staining system (Leica).
CEACAM6 expression in human leukocytes (monocytes, neutrophils and T cells) from whole blood of healthy volunteers (CRS Clinical Research Services Berlin GmbH, Berlin, Germany) and on human dissociated tumor cells and infiltrating neutrophils, macrophages and T cells from five colorectal cancer (CRC) and seven NSCLC patients (Conversant Bio, Inc. USA) was examined by flow cytometry. To analyze CEACAM6 expression in different cell populations of the CRC and NSCLC samples, the cells were analyzed by gating on cell-specific surface markers CD45 (for leukocytes), CD14 (for monocytes and macrophages), CD3 (for T cells) and CEACAM8 (for granulocytes). The mAb 9A6 was used to assess CEACAM6 expression. The details of the analysis are described in the Supplementary Methods.
CEACAM6 RNA expression in various cancers
CEACAM6 mRNA expression was analyzed in primary tumor samples across 33 cancer types using The Cancer Genome Atlas (TCGA) database as described in the Supplementary Methods.
CEACAM6 silencing in various liquid and solid cancer cells by siRNA knockdown
Effects of CEACAM6 silencing were examined by small interfering RNA (siRNA) knockdown and co-cultures of TILs and cancer cells. PANC-1-luc cells were generated by transfecting PANC-1 cells with a plasmid encoding the green fluorescent protein (GFP)-luciferase fusion protein (pEGFP-Luc plasmid) and the G418-resistance gene. The plasmid was kindly provided by Dr. Haase (LMU-Munich, Munich, Germany). The protein expression and mRNA levels of PD-L1 and CEACAM6 were determined by Western blot and reverse-transcription PCR, respectively.
Wild-type and luc-transfected PANC-1 (2 x 103), KMM-1 (1 x 104), and NCI-H23 (2 x 103) cells were transfected with siRNAs (Suppl. Table S3) with RNAiMAX (Thermo Scientific). The cytotoxicity of the siRNAs was measured by a luciferase-based assay and a chromium-51 assay.
The details of the protocols are described in the Supplementary Methods.
Selectivity and binding of BAY 1834942
The binding and selectivity of BAY 1834942 to recombinant human CEACAM1, −3, −5 and −6 (R&D Systems) and cynomolgus monkey CEACAM6 were determined with surface plasmon resonance (SPR) and enzyme-linked immunosorbent assay (ELISA) experiments as described in the Supplementary Methods.
Additionally, binding and selectivity were examined in HeLa cells transfected with human CEACAM1, −3, −5, −6, −8, −19) or cynomolgus monkey CEACAM6 receptors by flow cytometry. Wild-type and stably transfected HeLa cells were incubated with BAY 1834942 as primary antibody and with 9A6 as a positive control. Phycoerythrin (PE)-labeled anti-mouse or anti-human IgG antibodies (1:150 dilution, #115–115-164, #109–115-098, Dianova) were used as secondary antibodies. For the analysis of half-maximal binding concentration (EC50), the primary antibodies were used at increasing concentrations of 0.1 nM to 100 nM. EC50 values were determined by plotting the median fluorescence intensity signal against the concentration (logarithmic scale). Curve fitting of data was performed using GraphPad Prism 6.0. The details are described in the flow cytometry section of the Supplementary Methods
The interaction between recombinant CEACAM6 and CEACAM1 Fc fusion protein was tested by ELISA. CEACAM6-Fc (0.039–0.5 µg/mL) was added to CEACAM1-coated (without Fc, R&D #2244) microtiter plates for 1 h at 37°C. Detection was performed with a secondary antibody (1:5000 goat anti-human IgG horseradish peroxidase, DIANOVA #109–035_098) for 1 h at 37°C and developed after incubation with substrate (R&D #DY999 A and B) and stopping solution (R&D #DY994). Plates were measured at 450/540 nm with a Tecan Infinite M200 plate reader. Analysis was performed using Microsoft Excel and Graph Pad Prism 6. The details are described in the Supplementary Methods.
The effect of BAY 1834942 on CEACAM6-CEACAM1 interaction was analyzed in a similar setup as a competition ELISA using a range of BAY 1834942 concentrations. Therefore, a precomplex of CEACAM6-Fc (2 µg/mL) and increasing concentrations of BAY 1834942 was allowed to form at 37°C for 1 h before it was added to the CEACAM1-coated plates.
To analyze the binding of BAY 1834942 to human CEACAM6 in more detail, two mutants of the N-terminal domain 1 of human CEACAM6 were generated with changes at amino acid position 30 (Suppl. Table S4) and investigated alongside the wild-type domain. The details of the experiment are described in the Supplementary Methods.
Flow cytometry analyses for profiling of cancer cell lines, T cells, or TILs
The expression of various markers (e.g., CEACAM6, CEACAM5, CEACAM1, and PD-L1) on the surface of cancer cell lines, survivin T cells, virus-specific T cells and pancreatic cancer TILs was analyzed using flow cytometry on a FACS Array (BD) or FACSCanto II instrument (BD). Data were analyzed with FlowJo (V10.2) software (Tree Star). Determination of antibody binding sites on individual cell lines was done with the QIFI® kit (Dako) according to the manufacturer’s instructions. Method details can be found in the Supplementary Methods.
Effect of BAY 1834942 on cytokine release from co-cultures with potential target cancer cell lines
The efficacy of BAY 1834942 on T cell activity was assessed in co-culture assays. Survivin T cells, naïve PBMCs from healthy donors, or pancreatic cancer TILs were co-cultured with various CEACAM6-expressing or CEACAM6-negative cancer cells, and the subsequent levels of secreted cytokines (e.g. interferon-γ (IFN-γ), IL-2, and tumor necrosis factor-α (TNF-α)) were measured by IFN-γ ELISpot, sandwich ELISA, or Luminex multiplex analyses as described in the Supplementary Methods.
Analysis of T cell-mediated tumor cell killing
T cell-mediated tumor cell killing was analyzed in co-cultures of lung cancer cells with TILs in the presence of BAY 1834942 using an impedance-based xCELLigence cytotoxicity assay according to manufacturer’s instructions (RTCA Device, ACEA Biosciences). Data were analyzed using RTCA Data Analysis software 2.0, Microsoft Excel 2010 and GraphPad Prism 6.
Effects of BAY 1834943 on cytokine secretion in combination with immune checkpoint molecule antibodies or CEACAM1 antibody
The efficacy of BAY 1834942 in combination with an antibody directed against CEACAM1 or antibodies inhibiting immune checkpoint molecules PD-1 or TIM-3 was studied in co-culture assays of 10,000 HCC2935 lung cancer cells with 20,000 FluM1 peptide-specific T cells. The virus-specific T cells were stimulated with the associated FluM1 peptide at 0.2 μg/mL. Anti-TIM-3 was used at 50 µg/mL, and all other antibodies at 30 µg/mL. In the combination experiments, BAY 1834942 was applied approximately at its EC50 of 1 µg/mL to ensure the effects of other antibodies on the activation of T cells. The co-cultures were incubated at 37°C, 5% CO2 for approximately 20 h and the IFN-γ cytokine secretion was measured in the supernatants by sandwich ELISA.
The virus peptide antigen specificity of the virus-specific T cells was confirmed with tetramer (F391–4A-E, ProImmune) staining and flow cytometry analysis before the co-culture experiments.
Effect of CEACAM6, CEACAM1 and BAY 1834942 on T cell signaling
The effects of CEACAM6 and CEACAM1 on T cell signaling were studied in CD3-preactivated human Jurkat T lymphocytes by immunoprecipitation (IP) and Western blot using zeta-chain-associated protein kinase 70 (ZAP70) (Cell Signaling) and pZAP70 rabbit antibodies (Cell Signaling) and donkey anti-rabbit antibodies (R&D Jackson Immunoresearch) for detection as described in the Supplementary Methods.
For immunosuppression, Jurkat cells were incubated for 2 min with beads loaded with recombinant human CEACAM6 or -1 or huIgG1 Fc in the presence of anti-CD3 antibody (OKT3, eBiosciences) before they were lysed and cleared for IP.
The role of CEACAM1 in BAY 1834942-induced effects on T cell activation (IL-2 secretion) was examined in a co-culture of 100,000 CEACAM1 knockout or wild-type Jurkat T cells (preactivated with anti-CD3 OKT3, 0.5 µg/mL, plate-bound, at 37°C for 90 min) and 50,000 HCC2935 lung cancer cells in the presence of BAY 1834942 (40 µg/mL in X–vivo 20) or isotype control antibody and the bispecific epithelial cellular adhesion molecule (EpCAM)/CD3 antibody (10 ng/mL). IL-2 cytokine levels in supernatants were measured by ELISA. The Jurkat T cell CEACAM1 knockout generation has been described in the Supplementary Methods.
The ability of BAY 1834942 and of an anti-CEACAM1 antibody to reconstitute T cell activity was assessed by measuring IFN-γ cytokine levels in survivin T cells that had been pre-suppressed with beads loaded with CD3 antibody and recombinant human CEACAM6 and incubated for 20 h. Beads loaded with anti-CD3 antibody and huIgG1 Fc (Eureka) were used as a control. Details are described in the Supplementary Methods.
Statistics
Unless stated otherwise, all analyses were performed using the statistical programming language R (version 3.4.4). Validity of the model assumptions was checked for each fitted statistical model. All analyses, unless stated otherwise, were performed using a linear model estimated with generalized least squares that included separate variance parameters for each study group. In addition to the treatment effect, the models included a fixed effect for each sample. Mean comparisons between the treatment groups were performed using the estimated linear model and corrected for family-wise error rate using Sidak’s method.
Exceptions to this pattern were the analyses of SW1116 and AsPC-1 co-culture assays, where separate models were fitted for anti-CD19 and anti-EpCAM data; as well as the analysis of cytokine release over the course of time, where each timepoint was treated as a continuous covariate resulting in linear regression lines to be fitted for each treatment group. Comparisons at different time points are based on this fitted model rather than only the data collected at those time points.
Synergy was investigated using a linear model estimated using weighted least squares that had a separate variance term for each group. 0 was used as the reference level for treatment effects.
Results
High CEACAM6 expression in colorectal, pancreatic, gastric and lung cancer
We analyzed CEACAM6 mRNA expression using the TCGA database (Suppl. Fig. S1A). CEACAM6 expression was high in over 75% of CRCs, NSCLCs, and pancreatic cancers. In these entities, over 50% of the samples exhibited the highest expression, defined as >600 transcripts per million (TPM). Accordingly, a systematic comparative IHC analysis of 697 primary tumors revealed CEACAM6 expression at the highest frequency and intensity in CRC, NSCLC, gastric, gastroesophageal junction (G-E junction), and pancreatic cancers ()). A semi-quantitative immunohistochemical analysis of CEACAM6 and PD-L1 expression in NSCLC samples showed no correlation between CEACAM6 and PD-L1 expression (), Suppl. Fig. S1B).
Figure 1. Expression of CEACAM6 in cancerous tissues and blood cells. A. CEACAM6 expression in tissue sections of solid tumors as detected by immunohistochemistry with mAb 9A6. CRC, colorectal carcinoma; NSCLC, non-small cell lung adenocarcinoma. B. Prevalence (%) of CEACAM6 expression in various cancer types as categorized by H-scores. H-score was calculated as 3 x (% tumor cells of staining intensity 3) + 2 x (% tumor cells of staining intensity 2) + 1 x (% tumor cells of staining intensity 1), range 0–300. ADC, adenocarcinoma; G-E, gastroesophageal; HNSCC, head and neck squamous cell carcinoma; RCC; renal cell carcinoma; SCC, squamous cell carcinoma. C. Comparison of CEACAM6 and PD-L1 expression in NSCLC (n = 50) as determined by H-scores. Using a 5% positivity cutoff for both parameters (CEACAM6-positive ≥ 5%; PD-L1-negative ≤ 5%), 26% of tumor samples can be considered positive for both targets. D. Representative histograms of CEACAM6 expression on dissociated CRC (samples #1 and #3) and NSCLC (samples #2 and #4) tumor cells as detected by flow cytometry using a phycoerythrin (PE)-labeled anti-CEACAM6 antibody 9A6. Cells were gated according to cell-specific surface markers: epithelial cellular adhesion molecule (EpCAM, clone HEA-125, Miltenyi #130–098–113) for tumor cells), CD45 (clone H130, BioLegend, #304036) for leukocytes, CD14 (clone M5E2, BioLegend, #301830) for monocytes and macrophages, CD3 (clone SK7, BD Pharmingen, #560275) for T cells, and CEACAM8 (CD66b, clone G10F5, BioLegend #305107) for granulocytes.
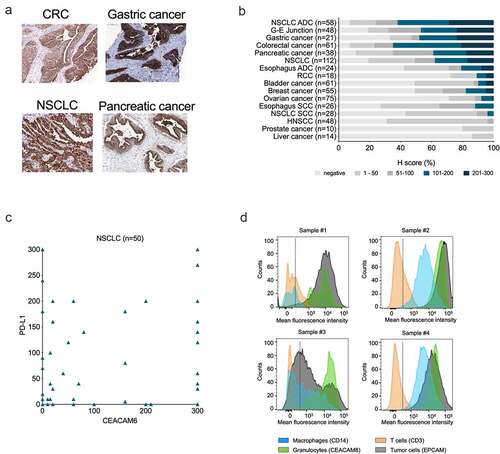
The IHC analysis of normal tissues confirmed CEACAM6 expression in the lung and colon, and on resident myeloid cells in the spleen, lymph nodes, and small intestine (Suppl. Fig. S1C). In the blood, CEACAM6 was expressed on granulocytes and weakly on monocytes while CEACAM6 staining on T cells did not reveal significant differences compared to negative controls (Suppl. Fig. S1D).
Flow cytometric analysis of dissociated cells from CRC and NSCLC tumors showed high variability in CEACAM6 expression levels on cancer cells, while the levels were consistently high in all samples on tumor-infiltrating granulocytes and, to a lesser extent, on macrophages ()). No notable CEACAM6 expression was detected on T cells in any of the samples. It should be noted that in sample #1, a heterogeneous stain of CEACAM6 on neutrophilic granulocytes was observed, which may indicate differences in the activation state, as CEACAM6 is known to be upregulated upon activation of granulocytes.Citation40
CEACAM6 mediates immune suppression in solid tumors
To assess whether not only malignant plasma cells but also epithelial cancer cells use CEACAM6 to inhibit T cell activity, HLA-A2-positive, luciferase-transfected human cancer cells NCI-H23 (NSCLC) and PANC-1 (pancreatic cancer) were studied in conjunction with cytotoxic T cells and compared with luciferase-transfected malignant KMM1 plasma cells. The functionality of this reporter system was confirmed by reduced luciferase activity in transfected PANC-1-luc cells upon siRNA-mediated silencing of cell viability genes or upon knockdown of luciferase expression ()).
Figure 2. CEACAM6 expression in tumor cells abrogates cytolytic capacity of tumor-infiltrating lymphocytes (TILs). 9A6 was used as the CEACAM6-specific antibody and scrambled, nonspecific RNA (scr) in (a-g) and an isotype control antibody in (h) were used as controls. A. Reduction of luciferase signal upon transfection with a mixture of siRNAs inducing cell death (siCD), ubiquitin C (UBC, a gene essential for cell survival), and firefly luciferase (siFLuc) in PANC-1-luc pancreatic cancer cells. Analysis was performed by a luciferase-based viability assay. B.-D. Effect of siRNA knockdown of CEACAM6 on T cell-mediated killing of (b) KMM-1-luc multiple myeloma, (c) PANC-1-luc, and (d) NCI-H23-luc NSCLC cancer cells. Cells were co-cultured with or without matched cytotoxic TILs and cancer cell survival was determined by measuring the remaining luciferase activity after 20 h. E.-F. Effect of siRNA knockdown of CEACAM6, PD-L1, RCAS-1, or GAL-3 on T cell-mediated killing of (e) NCI-H23-luc and (f) PANC-1-luc cancer cells. The experiment was conducted as in (b-d). G. Effect of siRNA knockdown of CEACAM6 or PD-L1 on T cell-mediated killing of PANC-1 cancer cells after a 6-h co-culture as determined by a chromium release assay. H. Effect of CEACAM6 blocking by anti-CEACAM6 antibody 9A6 on T cell-mediated killing of PANC-1 cancer cells after a 6-h co-culture as determined by a chromium release assay. Untreated wild-type PANC-1 cells and an isotype antibody were used as controls. Graphs show representative data of at least two independent experiments. Statistical analyses were performed using a linear model using weighted least squares that a had separate variance term for each group (a, b, d, e); ANOVA with homogeneous variance across all groups (c); pair-wise comparisons using Welch’s t-test and Bonferroni correction (f); or linear regression using log2 of ratio as a covariate (g, h). Stars denote statistically significant differences in comparison to control or vehicle. ***, p < .001. NSCLC, non-small cell lung carcinoma; RLU, relative light unit; siRNA, small interfering RNA; TIL, tumor-infiltrating lymphocyte.
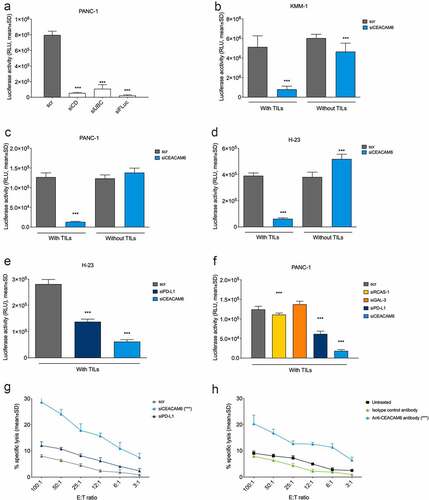
The cancer cell lines were co-cultured with TILs derived from HLA-A2-matched patients with the corresponding cancer. The TIL populations from pancreatic cancer used in this study were already reported earlier.Citation41 The CD4/CD8 composition of these TILs and of those derived from lung cancer and multiple myeloma are shown in Suppl. Fig. S2. All TIL cultures were highly enriched for T cells (>90%) and did not contain considerable amounts of monocytes, macrophages, or granulocytes. The maximum proportion of Treg cells observed was 5.3% (data not shown).
CEACAM6 knockdown induced efficient cancer cell killing by TILs to an equal extent in both multiple myeloma (KMM-1) and solid cancer (NCI-H23 and PANC-1) cells, whereas no T cell killing (in KMM-1) or only low amounts of it (in NCI-H23 and PANC-1) were observed in the mock-transduced cells ()). When comparing CEACAM6 to established ICMs, CEACAM6 knockdown was more effective in inducing T cell-mediated cancer cell lysis even in cells that showed robust expression of PD-L1 (Suppl. Fig. S3) than the knockdown of PD-L1, RCAS-1, or GAL-3 ()).
Analysis of chromium-51 release in dying PANC-1 cells confirmed that CEACAM6 silencing by siRNA resulted in a stronger T cell-mediated cell lysis than PD-L1 silencing ()). Notably, a similar increase in chromium-51 release was also achieved by blocking CEACAM6 on the surface of PANC-1 cells by mAb 9A6 ()), suggesting that surface-located CEACAM6 mediates immune inhibition and represents a potentially therapeutic target.
BAY 1834942 is a humanized, CEACAM6-selective antibody blocking CEACAM6 interaction with CEACAM1
BAY 1834942, a humanized IgG2 isotype mAb against human CEACAM6, was generated to block the immune checkpoint function of CEACAM6. In contrast to murine anti-human CEACAM6-specific mAb (9A6), BAY 1834942 binds to human and cynomolgus monkey CEACAM6 with monovalent dissociation constants of KD of 13 nM and 31 nM, respectively, as determined by an SPR analysis using recombinant extracellular domains (), Suppl. Table S5) and ELISA (), Suppl. Table S6). This binding profile was confirmed by flow cytometry in CEACAM6-transfected HeLa cell lines with full-length CEACAM6 variants. BAY 1834942 did not bind to wild-type HeLa cells (), Suppl. Fig. S4, Suppl. Table S7). The half-maximal binding EC50 values for BAY 1834942 and for the positive control antibody 9A6 (with modified human IgG2 backbone) were determined by flow cytometry in a panel of CEACAM6-expressing cancer cell lines which show binding to authentic CEACAM6 on cancer cells ()). For BAY 1834942, the EC50 values were in the range of 0.15 nM (PA-TU-8902) to 5 nM (HCC2935), which were comparable to 9A6 (0.2 nM to 6 nM). The differences in EC50 of BAY 1834942 and 9A6 and in between different cell lines tested may be due to different glycosylation patterns or the amount of CEACAM1 and CEACAM5 co-expressed in the cell lines.
Figure 3. Antibody-binding profile of BAY 1834942 as determined by ELISA and flow cytometry. A. Binding of BAY 1834942 to recombinant human and cynomolgus CEACAM6 that were coated on a CM5 sensor chip and analyzed by SPR. B. Selective binding of BAY 1834942 to recombinant human CEACAM6 and cynomolgus monkey CEACAM6 with absence of reactivity to human CEACAM1, −3, or −5 as determined by binding ELISA. C-D. Binding of BAY 1834942 and of a control antibody 9A6 (as huIgG2) to HeLa cells transfected with (b) human and (c) cynomolgus monkey CEACAM6 as determined by flow cytometry. No binding of BAY 1834942 was observed on HeLa wild-type cells. E. The half-maximal binding (EC50) values for BAY 1834942 and the positive control antibody 9A6 as determined by binding to CEACAM6-positive cancer cells by flow cytometry. F. Dose-dependent binding of recombinant CEACAM6-Fc to recombinant human CEACAM1 (coated to plate, 1 µg/mL) by ELISA. Human IgG antibody as Fc-control is shown on the right. G. BAY 1834942 and 9A6, added as precomplex of recombinant CEACAM6-Fc (2 µg/mL) and antibody in concentration series from 0.01–100 nM at 25°C for 1 h, interrupted the interaction between CEACAM1 (coated to plate, 1 µg/mL) and recombinant CEACAM6-Fc as determined in a competition ELISA assay. For detection of bound CEACAM6-Fc in the presence of BAY 1834942, an anti-human IgG horseradish peroxidase (HRP) conjugate was used with Amplex Red (Life Technologies) as substrate. RFU, relative fluorescence unit.
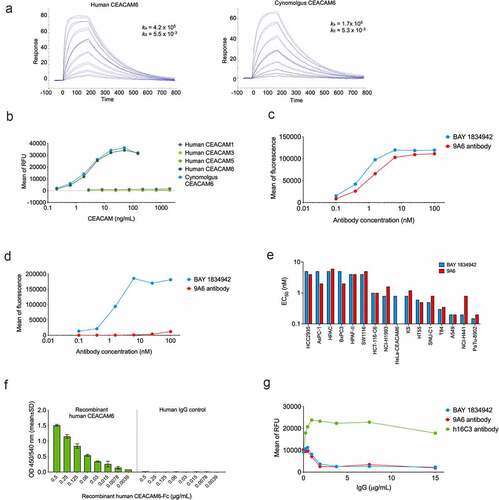
BAY 1834942 was highly selective for human CEACAM6, as it was unable to bind to human CEACAM1, CEACAM3, or CEACAM5 (), Suppl. Table S6). This was unexpected as the orthologous cynomolgus monkey CEACAM6 has 81% sequence identity with human CEACAM6 in its N-terminal domain, whereas the human paralogs CEACAM1, -3, or -5 show higher sequence identities with CEACAM6 (90%, 90%, and 89%, respectively). We studied this further by mutational analysis and found that BAY 1834942 binds to the N-terminal domain 1 of CEACAM6 even when wild-type Ile30 is replaced by Leu (mimicking the respective residue at this position in cynomolgus monkey CEACAM6). However, the mutation of Ile30 to Phe (the respective residue in human CEACAM1, -3, and -5) completely abolished the binding (Suppl. Fig. S5). This indicates that BAY 1834942 exploits position 30 to a large extent to gain its selectivity and cross-reactivity profile.
We also investigated the potential interaction between CEACAM6 and CEACAM1 on T cells. Binding of recombinant CEACAM6-Fc to CEACAM1-coated plates was blocked by BAY 1834942 in a dose-dependent manner ()). In contrast, the antibody h16C3, earlier reported to bind CEACAM5/6,Citation35,Citation36 was found to be unable to interrupt the interaction of CEACAM1 and CEACAM6 ()).
BAY 1834942 increases type 1 cytokine secretion and the cytotoxic capacity of tumor-specific T cells
The effect of CEACAM6 blockade on T cell activation was analyzed in co-cultures of CEACAM6-expressing tumor cells and tumor-specific T cells. BAY 1834942 increased the secretion of IFN-γ over 1.5-fold in co-cultures of T cells recognizing the tumor antigen survivin (survivin T cells) with various survivin-expressing cancer cells. These cancer cells included KS22.24 breast cancer, HCC2935 lung cancer, HPAC, HPAFII, and AsPC-1 pancreatic cancer, as well as CEACAM6-transfected HCT-116 colon cancer cells (, Suppl. Fig. S6A–C, ). The magnitude of the effects correlated with the CEACAM6 expression levels on the cancer cells (–e), ) and did not correlate with co-expressed CEACAM1 or CEACAM5 on the same cell lines (Suppl. Table S8). In these settings, the efficacy (EC50) of BAY 1834942 to induce cytokine release was in the range of 0.28–3.4 nM (, Suppl. Table S9), which was comparable to the EC50 values (0.15–5 nM) determined for CEACAM6-positive cells ()).
Figure 4. Immunosuppression and efficacy of BAY 1834942 correlates with receptor density and is dose dependent. A-D. BAY 1834942-induced (30 µg/mL) increase in IFN-γ cytokine release in co-culture assays of 10,000 (a) SNU-C1 colon, (b) NCI-H1993 lung, (c) KS22.24 breast, or (d) CEACAM6-transfected HCT-116 colon cancer cells and 20,000 survivin T cells. Representative examples are shown in the figure. Full data set shown in . IFN-γ levels were measured by ELISA and fold change in the induction of IFN-γ levels over background is indicated with an arrow. The data were normalized to the mean of the isotype control-treated group. E. CEACAM6 expression in SNU-C1, NCI-H1993, KS22.24, and CEACAM6-transfected HCT-116 cells as analyzed by flow cytometry and presented as mean fluorescence intensity (MFI) after binding of CEACAM6 mAb (murine IgG2a variant of BAY 1834942 for use with the Qifi kit (Dako)). Full data set shown in . F.-H. Examples of dose-dependent cytokine secretion in co-culture of 10,000 (f) KS breast cancer, (g) HCC2935 lung cancer, and (h) CEACAM6-transfected HCT116 colon cancer cells with 20,000 survivin T cells. BAY 1834942 and isotype control antibody were used at concentrations ranging from 0.6 ng/mL to 30 µg/mL. IFN-γ levels were measured with ELISA. I.-J. Effect of BAY 1834942 on the killing of HCC2935 lung cancer cells by TILs isolated from pancreatic cancer patients. EpCAM/CD3 bispecific antibody construct (0.25 ng/mL) was used as a T cell-engaging molecule and tumor cell killing was measured by loss of impedance with an Xcelligence instrument. (i) Co-culture of 10,000 HCC2935 tumor cells and 50,000 TILs (TIL-12) with BAY 1834942, 9A6 anti-CEACAM6 mAb (huIgG2 variant), anti-PD-L1 antibody (huIgG2 variant) and isotype control used at 30 µg/mL. (j). Same co-culture as in (i) with BAY 1834942 used at doses 0.02–50 µg/mL and isotype control antibody (50 µg/mL) as a control. Statistical analysis in (a-d) was performed in comparison to the isotype control group using a linear model and corrected for family-wise error rate using Sidak’s method. ***, p < .001.
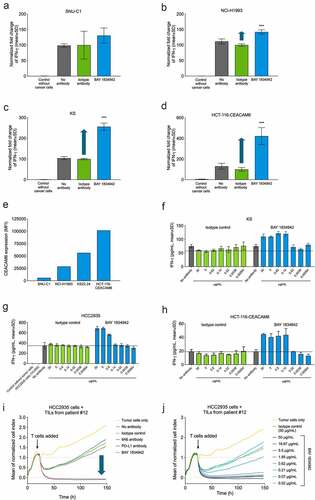
Table 1. Correlation of BAY 1834942 induced IFN-γ secretion with CEACAM6 expression in various human cancer cells, as determined by cytokine release assays on co-cultures with cancer cells (10,000 cells) and survivin T cells (20,000 cells)
Further analysis revealed increased secretion of cytokines IFN-γ, IL-2, and TNF-α in co-cultures of survivin T cells and KS22.24 breast cancer cells (Suppl. Fig. S6D). When IFN-γ secretion was measured in co-cultures of HCC2935 lung cancer cells over time, cytokine secretion occurred rapidly within 24 h and was sustained for up to 96 h (Suppl. Fig. S6E). These results were further supported by similar and dose-dependent observations in co-cultures of patient-derived TILs and HCC-2935 lung cancer cells (Suppl. Fig. S6F–G) and co-cultures of survivin T cells and CEACAM6-transfected HCT116 colon cancer cells (Suppl. Fig. S6H).
In addition to cytokine secretion, BAY 1834942 also increased T cell-mediated cytotoxicity dose-dependently in a co-culture of TILs and HCC2935 lung cancer cells with high CEACAM6 expression ()). PD-L1 blockade showed no effect on tumor cell killing in this setting, possibly due to weak PD-1 expression in the employed TILs or because of the dominance of the immunosuppressive effect of CEACAM6 ()). Similar results were obtained with a second TIL preparation (Suppl. Fig. S6I–L).
Immunosuppression by CEACAM6 is mediated via CEACAM1 on activated T cells
The expression of different CEACAM receptors on activated T cells was studied to identify potential candidate receptors for CEACAM6. CEACAM1 was the only CEACAM receptor family member expressed on activated T cells ()). Accordingly, combining BAY 1834942 with an anti-CEACAM1 antibody to block both CEACAM6 and CEACAM1 did not result in any further increase of IFN-γ secretion in a co-culture of virus-specific T cells and HCC2935 lung cancer cells ()). Furthermore, the effect of CEACAM6 blockade by BAY 1834942 was dependent on the presence of CEACAM1 on T cells (Suppl. Fig. S7). We determined this by comparing the IL-2 secretion from activated, CEACAM1-expressing wild-type Jurkat T cells (Suppl. Fig. S8) with the secretion from CEACAM1 knockout Jurkat T cells co-cultured with HCC2935 lung cancer cells ()). Together, these data suggest that CEACAM6 and CEACAM1 (inter)act on the same axis and support the hypothesis that CEACAM1 on activated T cells is a receptor for CEACAM6. Indeed, both recombinant human CEACAM6 and CEACAM1 suppressed CD3-induced T cell receptor signaling of Jurkat T cells, as indicated by reduced levels of pZAP70, determined by IP and Western blot ()). Blockade of CEACAM6 by BAY 1834942 restored T cell activity in a dose-dependent manner, as evidenced by increased IFN-γ secretion by survivin T cells co-incubated with recombinant CEACAM6-loaded beads ()). Importantly, immunosuppression by recombinant CEACAM6-loaded beads was completely removed by antibody blockade of CEACAM1 on T cells, confirming that CEACAM1 is the only relevant inhibitory receptor mediating the immunosuppressive effects of CEACAM6 on T cells ()).
Figure 5. Immunomodulatory characteristics of BAY 1834942. A. Analysis of CEACAM1, -5, -6, and -8 receptor expression on survivin T cells and virus-specific T cells by flow cytometry using anti-CEACAM1 (TPP-3006) and anti-CEACAM6 (BAY 1834942). All other antibodies as listed in Suppl. Table S1. B. Effects of combined antibody-mediated blockade of CEACAM6 and CEACAM1 on IFN-γ cytokine secretion. Co-culture of 10,000 FluM1 peptide-loaded HCC2935 cancer cells with 20,000 FluM1-specific T cells for 20 h. BAY 1834942 and CEACAM1 antibody (TPP-3006) were used at 30 µg/mL. In combination experiments, BAY 1834942 and isotype mAbs were used at 1 µg/mL. IFN-γ levels were measured by ELISA. The isotype control antibodies (30 µg/mL) used were TPP-1238 (huIgG2 isotype control) and TPP-754 (huIgG1 isotype) control. The percentages indicate the fold differences in cytokine secretion. C. The effect of BAY 1834942 on IL-2 secretion in 100,000 CEACAM1 knockout Jurkat E6.1 or wild-type Jurkat E6.1 cells (both CD3-preactivated) after a 20-hour co-culture with 50,000 HCC2935 lung cancer cells in the presence of T cell engaging antibody construct EpCAM/CD3 (10 ng/mL) and either BAY 1834942 (40 µg/mL in X–vivo 20 medium) or TPP-1238 isotype control antibody. IL-2 levels in supernatants were analyzed by ELISA. The percentages indicate the fold differences in cytokine secretion. D. Recombinant CEACAM6 and CEACAM1 loaded on beads suppressed T cell signaling as determined from phosphorylated zeta-chain-associated protein kinase 70 (pZAP70, Tyr319) levels in CD3-activated Jurkat T cell lysates by immunoprecipitation and Western blot. HuIgG1 mAb Eureka was used as the Fc-isotype control. E. CEACAM6-suppressed cytokine secretion was reconstituted by BAY 1834942 in survivin T cells (100,000 cells) co-incubated for 20 h with 1 × 106 CEACAM6-loaded beads as determined by IFN-γ levels measured with ELISA. The CD3/CEACAM6 beads were pre-incubated with BAY 1834942 or an isotype control antibody at room temperature for 20 min. The colored closed circles denote reactions as follows: Black circles: beads coated with huIgG1; Yellow circles: beads coated with anti-CD3 and huIgG1; Orange circles: beads coated with anti-CD3, huIgG1, and CEACAM6-Fc; Green circles: beads coated with anti-CD3, huIgG1, and CEACAM6-Fc + increasing concentrations of isotype control TPP-1238 in solution; Blue circles: beads coated with anti-CD3, huIgG1, and CEACAM6-Fc + increasing concentrations of BAY 1834942 in solution. F. Reversal of CEACAM6-induced suppression of cytokine secretion as described in (e) using an anti-CEACAM1 (TPP-9145) antibody and an isotype control. The colored closed circles denote reactions as follows: Black circles: beads coated with huIgG1; Yellow circles: beads coated with anti-CD3 and huIgG1; Orange circles: beads coated with anti-CD3, huIgG1, and CEACAM6-Fc; Green circles: beads coated with anti-CD3, huIgG1, and CEACAM6-Fc + increasing concentrations of isotype control TPP-754 in solution; Blue circles: beads coated with anti-CD3, huIgG1, and CEACAM6-Fc + increasing concentrations of the anti-CEACAM1 antibody (TPP-9145) in solution. The anti-CEACAM1 antibody was pre-incubated with Jurkat cells; Pink circles: beads coated with anti-CD3 and huIgG1 + anti-CEACAM1 antibody (TPP-9145) in solution; Dark purple circles: beads coated with huIgG1 + anti-CEACAM1 antibody (TPP-9145) in solution. Statistical analysis for (b-c) was performed using a linear model and corrected for family-wise error rate using Sidak’s method. ns, non-significant; *, p < .05; ***, p < .001.
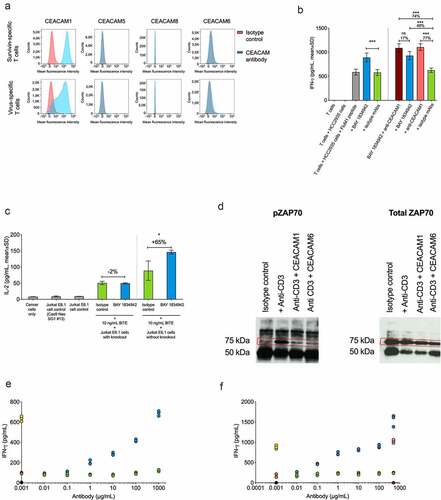
We therefore confirmed CEACAM1 expression in human cultured or freshly isolated CD4 and CD8 T cells from various sources such as cultured PBMCs and TILs by flow cytometry, and by reanalysis of published single cell RNA data sets from malignant melanomas and lung cancers.Citation42,Citation43 CEACAM1 was expressed on all CD4 and CD8 T cells in in vitro-expanded TIL cultures. PBMCs showed comparable CEACAM1 induction on both CD4 and CD8 T cells 72 h after polyclonal stimulation at levels ranging between 5–40% (Suppl. Fig. S9A). High CEACAM1 expression was also found in largely pure CD8 T cell cultures such as a survivin-specific CD8 T cell clone (Suppl. Fig. S9B). CEACAM1 mRNA-expressing T cells were present in 19/22 (86%) of melanoma samples and in 14/14 (100%) of lung cancer samples. Their proportions ranged between 0–30% for CD8 and 0–45% for CD4 in CEACAM1-expressing cells. On average, 5–17% of the respective T cell subsets showed CEACAM1 mRNA expression. In general, CEACAM1 expression was higher in CD4 TILs from both cancer types, but considerable proportions of CEACAM1-expressing cells were also found in the CD8 compartment (Suppl. Fig. S9C).
BAY 1834942 shows equal or higher efficacy compared to the anti-PD-L1 antibody
We next compared the efficacy of BAY 1834942 to PD-L1 inhibition in vitro in four different TIL cultures. In a flow cytometric analysis, all of these TILs showed CEACAM1 expression but low PD1 expression ()). In co-cultures of TILs from patients #31 and #34 and CEACAM6- and PD-L1-double-positive HCC2935 lung cancer cells, BAY 1834942 increased IFN-γ secretion while anti-PD-L1 showed no effect in comparison to the isotype control ()). Comparable results were obtained with all four batches of patient-derived TILs tested and also when using another cytokine, granzyme B, as the readout (Suppl. Fig. S10A–E).
Figure 6. Effects of BAY 1834942 and anti-PD-1, anti-PD-L1 and anti-TIM-3 antibodies on IFN-γ cytokine secretion. Co-cultures of HCC-2935 lung cancer cells and tumor-infiltrating lymphocytes from pancreatic cancer patients were used in addition to combination experiments employing FluM1 virus-specific T cells. Antibodies were used at 30 µg/mL and EpCAM/CD3 bispecific antibody construct at 0.1 or 0.25 ng/mL as a T cell-engaging molecule. A.-B. Expression of CEACAM1 and PD-1 in TILs from (a) patient #31 and (b) patient #34 as determined by flow cytometry after gating on CD3-positive cells. The specific staining signals for anti-CEACAM1 (TPP-3006) and anti-PD1 are shown in blue and for the isotype controls in orange. C.-D. IFN-γ secretion (ELISA) from co-culture of 10,000 HCC2935 lung cancer cells and 20,000 TILs from (c) patient #31 and (d) patient #34. Statistical analysis was performed in comparison to the isotype control group using a linear model and corrected for familywise or false positive error rates using Dunnett’s or Sidak’s method. *, p < .01; ***, p < .001. E. Flow cytometry analysis of CEACAM6 and PD-L1 expression in HCC2935 lung cancer cells. The specific signals for BAY 1834942 and anti-PD-L1 are shown in blue and for the isotype control in red. F. Flow cytometry analysis of virus-specific T cells. Cells were gated on CD3 and CD8 and analyzed for CEACAM1 (TPP-3006), PD-1, and TIM-3 expression on day 14 after peptide induction. G.-H. Effects of BAY 1834942 on T cell activation in co-culture of CEACAM6-positive and PD-L1-positive FluM1-loaded HCC2935 lung cancer cells (10,000 cells) and virus-specific T cells (20,000 cells) in combination with inhibitory antibodies against checkpoint molecules (g) PD-1 (30 µg/mL), and (h) TIM-3 (50 µg/mL). For combination, BAY 1834942 was used at 1 µg/mL. Data represent mean percent increase in IFN-γ cytokine secretion (ELISA) by T cells as compared to respective isotype control antibodies. Each symbol represents an independent experiment with colors symbolizing the same co-cultures used. The median and SD are indicated with horizontal lines. Statistical analysis was performed using a linear model and corrected for family-wise error rate using Sidak’s method. *, p < .05; **, p < .01; ***, p < .001.
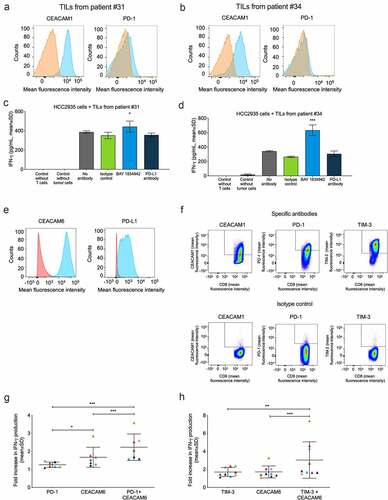
Enhanced efficacy with combination of BAY 1834942 and immune checkpoint inhibition
Finally, we tested the combination of BAY 1834942 with antibodies against ICMs in a PD-L1/PD-1-positive system consisting of virus-specific T cells and HCC2935 lung cancer cells that were loaded with the corresponding FluM1 virus peptide for T cell recognition. CEACAM6 and PD-L1 expression in HCC2935 cancer cells and the expression of ICM receptors CEACAM1, PD-1, and TIM-3 on virus-specific FluM1 T cells are shown in and Suppl. Fig. S11A. We detected significant, at least additive efficacy in terms of cytokine secretion and thus, T cell activation, when BAY 1834942 was combined with antibodies against PD-1 (p < .001) or TIM-3 (p < .001) (, Suppl. Fig. S11B).
Discussion
Here, we report on an immunosuppressive axis between CEACAM6 on epithelial tumor cells and CEACAM1 on tumor-reactive T cells and on its inhibition with the CEACAM6-specific antibody BAY 1834942.
Using in vitro analyses, we show that the CEACAM6/CEACAM1 interaction can serve as an immunosuppressive mechanism in the interaction between human activated cytotoxic T cells and epithelial cancer cells. It is independent of the PD-L1/PD-1 axis and is active in several tumor entities resistant to PD-1 blockade. We show that CEACAM6 suppresses antitumor responses such as cytokine secretion and T cell-mediated tumor killing in both PD-L1-positive and PD-L1-negative cancer cells and does this equally or more efficiently than PD-L1. While T cell inhibition by PD-L1 is mediated through PD-1, we show that T cell inhibition by CEACAM6 is fully mediated through CEACAM1. Thus, the extent of CEACAM6-mediated immunosuppression is co-determined by the extent of CEACAM1 expression on T cells. Since CEACAM1 is transported to the cell surface upon stimulation of T cell receptors,Citation11,Citation12,Citation44 it is likely that this axis acts particularly on the small population of tumor-reactive cytotoxic T cells in the tumor microenvironment.Citation45 An immunoinhibitory role for CEACAM1-CEACAM1 interactions through recruitment of SHP phosphatases which impair not only T cell receptor signaling but also TLR2-mediated immune cell activation,Citation46 has been well established. While we did not address the latter point in this study, we show that CEACAM6-CEACAM1 interactions impair the T cell receptor-related phosphorylation of ZAP70. Besides, intrinsic CEACAM1 expression on CD8 T cells in the absence of homo- or heterophilic in-trans interactions may also play a supportive role in stabilizing lck signaling, which is important for efficient T cell priming in situations of viral infections.Citation47 The dissection of stimulatory and regulatory roles of CEACAM1 presence and crosslinking on T cells warrants further investigations.
We detected a considerable proportion of human cancers with high expression of CEACAM6 but low PD-L1 expression. Thus, it is conceivable that these tumors exploit CEACAM6 in addition to, or instead of, PD-L1 to achieve tumor immune resistance. However, our analyses were solely based on ex vivo cultured T cells which expressed high levels of CEACAM1. Whether CEACAM1 expression in tumor-infiltrating T cells in situ is sufficient to efficiently suppress T cell activity upon CEACAM6 binding, represents an important question that still needs to be systematically resolved. So far, publicly available RNA expression analyses from tumor-infiltrating single cells in human primary melanoma and lung cancers have revealed the presence of CEACAM1-expressing T cells in the vast majority of these tumors at highly variable proportions reaching up to 30% (CD8 T cells) and 45% (CD4 T cells) in individual tumors. However, proportions of T cells expressing CEACAM1 isoforms reported to participate in T cell inhibition (CEACAM1L and CEACAM1S) may be lower. For example, a single cell mRNA analysis from CD4 breast cancer infiltrating T cells revealed altogether 13% CEACAM1-expressing cells, of which 32% expressed isoform L (which contains two ITIM motifs), 32% expressed isoform S (lacking ITIM motifs), and 36% expressed isoform C which represents a putatively secreted isoform of yet uncharacterized function.Citation48 Thus, CEACAM1 mRNA expression seems to be much lower than PD1 expression in TILs of these tumors and whether the observed expression levels per cell reach functional significance in situ remains to be elucidated. Therefore, our study does not provide proof of physiological relevance of CEACAM6-mediated immunosuppression during epithelial cancer progression.
In contrast to the high affinity interactions between other checkpoint inhibitors such as PD-1 and PD-L1, interactions of CEACAMs are of low affinity.Citation13 A potential explanation is that in the immune synapse, many CEACAM molecules can interact with each other at the same time. This hypothesis is in line with our observation that a high receptor density (higher than threshold) is required to suppress T cell activity effectively and may provide an alternative mechanism for checkpoint regulation in tissues. Future studies addressing CEACAM protein:protein interactions and subsequent signaling cascades in the immunological synapse are warranted to clarify this question. Interestingly, the interaction of CEACAM6 with other receptors including CEACAM1 was also found by others in peptide interaction assays.Citation49
Based on the mouse mAb 9A6 (as positive control), we generated de novo BAY 1834942, a humanized, CEACAM6-selective high-affinity antibody capable of blocking the interaction of CEACAM6 with CEACAM1. BAY 1834942 and 9A6 displayed slightly different EC50 values, e.g., with respect to NCI-H441 cells and in between the different cell lines tested, which may be due to differences in the glycosylation patterns or the amount of CEACAM1 and CEACAM5 co-expressed in these cell lines. BAY 1834942 recognizes a different domain in CEACAM6 than the other reported antibodies such as h16C3,Citation35,Citation36 and it represents the only human CEACAM6-specific antibody that cross-reacts with cynomolgus monkey CEACAM6, thus enabling preclinical development.
BAY 1834942 efficiently restores antitumor T cell activity against various human cancers with low or no PD-L1 expression, but it shows activity also against PD-L1 positive cancer cells to a comparable or even stronger degree than PD-L1 blockade, particularly in the case of low PD-1 expression on T cells. This finding, together with the observation of independent expression of CEACAM6 and PD-L1 in e.g. NSCLC, confirms that the CEACAM6/CEACAM1 axis is independent of the PD-1/PD-L1 axis and points to the possibility that patients may benefit from treatment with a CEACAM6 antibody, even in settings where PD-1/PD-L1 inhibition is not effective. Nevertheless, we observed at least additive effects with a combination treatment with BAY 1834042 and anti-PD-1 when T cells co-expressed both CEACAM1 and PD-1. The most pronounced effects were observed when combining BAY 1834942 with an antibody inhibiting the second-generation ICM TIM-3. Despite the obvious limitations of in vitro assay systems, these data suggest that the responses to other checkpoint inhibitors can be improved by anti-CEACAM6 blockade.
Interestingly, both CEACAM1 and PD-1 exploit a similar inhibitory signaling mode since both recruit SHP phosphatases to the T cell receptor complex resulting in dephosphorylation of downstream kinases.Citation7,Citation50 Thus, it is conceivable that the recruitment of either of them to the immune synapse may sufficiently suppress T cell receptor signaling, leading to the rejection of the immune response against a tumor. Potentially, this mechanism might contribute to the insufficient sensitivity to anti-PD-1 immunotherapy sometimes observed in human cancers.
It remains to be evaluated whether anti-CEACAM6 therapies may have advantages over CEACAM1-blocking approaches which are currently being evaluated in clinical trials.Citation51 Conceptually, CEACAM6 targeting may improve tumor selectivity and the side-effect profile, since i) CEACAM1 binds also to itself, CEACAM5, CEACAM8, and most likely to other CEACAM family members as well.Citation11 These CEACAMs are expressed by multiple cell types throughout the body and may protect the cells against autoimmune destructionCitation11 and ii) CEACAM6 is normally expressed on the apical side of epithelial cells where it is not accessible for interactions with autoreactive T cells. In contrast, its expression on cancer cells is dysregulated and thus, it can translocate into the immune synapse with tumor-specific T cells.Citation8 Since CEACAM6 can be co-expressed with other CEACAMs on tumor cells, it remains open whether CEACAM6 blockade by antibodies alone will be sufficient to restore antitumor immunity in cancer patients. At least in our in vitro analyses, CEACAM6 was the dominant immunosuppressive CEACAM, as antitumor T cell reactivity could be efficiently increased upon CEACAM6 blockade even in the case of co-expression of other CEACAMs. Besides the immunostimulatory effects obtained by BAY 1834942 reported in this study, this antibody might also affect other properties of tumor cells and their microenvironment as far as they are mediated by CEACAM6. These were not subject of the present study and may require appropriate in vivo models.
Further preclinical validation of the immunotherapeutic potential of CEACAM6 blockade will indeed require in vivo efficacy studies with CEACAM6 antibodies. Unfortunately, these have not been possible due to the lack of a murine homologue of CEACAM6. Although a human CEACAM6-transgenic mouse has been developed,Citation28 this cannot be used for studying the immunomodulatory function of CEACAM6 as we were not able to detect an interaction between human CEACAM6 and murine CEACAM1 in our experiments (data not shown). An alternative approach of transplanting human CEACAM6-transfected murine tumor cells into human CEACAM1 transgenic miceCitation47 might help to study tumor rejection in vivo, but it would not mimic the CEACAM6-expressing myeloid compartment in the tumor stroma. Other humanized mouse models, including CD34 stem cell transplantation models, still have major caveats, as important parameters that define tumor immune rejection – e.g., antigen presentation by murine antigen-presenting cells, availability and suitability of cytokines and chemokines, interaction of human immune cells with murine vasculature and others – are not well matched yet.Citation52 Therefore, the physiological relevance of stem cell-based models for a humanized model is still questioned.Citation53 Furthermore, these models are highly variable and donor dependent. Thus, these models were not yet suitable for our study. Therefore, further assessment of the effects of CEACAM6 blockade in non-human primate models will be necessary before proceeding to clinical evaluation of BAY 1834942.
Taken together, CEACAM6 is an immune checkpoint with high expression in various solid cancers. CEACAM6 suppresses the anticancer activity of T cells in vitro and tumor cell rejection through interaction with CEACAM1. Our results suggest a therapeutic potential for combining CEACAM6 blockade with other checkpoint inhibitors. BAY 1834942 can efficiently inhibit the CEACAM6/CEACAM1 interaction and thus these data warrant evaluation of its therapeutic potential in clinical studies.
ABBREVIATIONS
ADC, adenocarcinoma; AML, acute myeloid leukemia; cDNA, complementary DNA; CEA; carcinoembryonic antigen; CEACAM6, carcinoembryonic antigen-related cell adhesion molecule 6; ccRCC, clear cell renal cell carcinoma; CLM, complete lymphocyte medium; CRC, colorectal carcinoma; DAB, diaminobenzidine; DLBCL, diffuse large B-call lymphoma; EC50, half-maximal effective concentration; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; ELISA, enzyme-linked immunosorbent assay; EpCAM, epithelial cellular adhesion molecule; FCS, fetal calf serum; FITC, fluorescein isothiocyanate; G-E, gastroesophageal; GFP, green fluorescent protein; GPI, glycosylphosphatidylinositol; HNSCC, head and neck squamous cell carcinoma; HRP, horseradish peroxidase; ICM, immune checkpoint molecule; IFN, interferon; Ig, immunoglobulin; IHC, immunohistochemistry; IL, interleukin; IP, immunoprecipitation; ITIM, immunoreceptor tyrosine-based inhibitory motive; mAb, monoclonal antibody; MFI, mean fluorescence intensity; NCA, nonspecific cross-reacting antigen; NHS, N-hydroxysuccinimide; NSCLC, non-small cell lung adenocarcinoma; PBMC, peripheral blood mononuclear cell; PBS, phosphate-buffered saline; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; PE, phycoerythrin; REP, rapid expansion protocol; RFU, relative fluorescence unit; RLU, relative light unit; SCC, squamous cell carcinoma; scr, scramble control; sgRNA, single guide RNA; siRNA, small interfering RNA; SPR, surface plasmon resonance; RCC, renal cell carcinoma; TBS, tris-buffered saline; TCGA, The Cancer Genome Atlas; TIL, tumor-infiltrating lymphocyte; TIM-3, T cell immunoglobulin mucin-3; TMA, tissue microarray; TNF, tumor necrosis factor; TPM, transcripts per million; UCOE, ubiquitous chromatin-opening element; V, variable; ZAP70, zeta-chain-associated protein kinase 70.
Authors’ contributions:
Designing research studies: JWi, PB, JP, MT, BK, RO, HHB, FW, WD. Conducting experiments: JWi, JP, VV, AS, AR, HHB, FW, EMG, JWe, OvA, MLE, JMG, CF, MX, SS. Acquiring data: JWi, JP, VV, AS, AR, RC, HHB, FW, EMG, JWe, OvA, DS, MLE, JMG, MX, JS, AN-M, AH, SS. Analyzing data: JWi, JP, VV, AS, AR, RC, HHB, FW, EMG, JWe, MT, GB, OvA, DS, MLE, JMG, WD, MX, RL, JS, AN-M, AH, SS. Providing reagents: JWi, RO, MT, DS, CF. Writing the manuscript: JWi, PB, JP, MT.
Supplemental Material
Download MS Word (22 MB)Acknowledgments
The authors gratefully acknowledge all involved lab technicians for excellent technical assistance, namely Annette Merling, Sabrina Purr, Giovanni Mastrogiulio, Simone Jünger, Kirstin Seifert, Jana Waetzold, and Beatrice Ölmez. The team is also grateful to Holger Hess-Stumpp and Ruth Wellenreuther for effective alliance management, Frank Dittmer, Sandra Bruder, Simon Holton and Nick Lobrenz for antibody and protein technology support, Barbara Nicke for support with the generation of Jurkat E6.1 CEACAM1 Crispr knockout cell lines, and Sabine Hoff for helpful discussions. VV was supported by DFG grant SFB-TR-221. Aurexel Life Sciences Ltd. (www.aurexel.com) is thanked for editorial assistance in the preparation of this manuscript, funded by Bayer AG.
Disclosure statement
Competing interests: JP, MT, WDD, RC, GB, JMG, DS, and JWi are employees of Bayer AG. JP, MT, WDD, OvA, and JWi are stockholders of Bayer AG. JP, HHB, MT, EMG, RC, CF, JMG, DS, and JWi are authors of CEACAM6 patents. CF, MLE, OvA, and JWe are former employees of Bayer AG.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Horna P, Sotomayor EM. Cellular and molecular mechanisms of tumor-induced T-cell tolerance. Curr Cancer Drug Targets. 2007;7(1):41–17. doi:10.2174/156800907780006940.
- Pardoll DM. Immunology beats cancer: a blueprint for successful translation. Nat Immunol. 2012;13(12):1129–1132. doi:10.1038/ni.2392.
- Yang L, Carbone DP. Tumor-host immune interactions and dendritic cell dysfunction. Adv Cancer Res. 2004;92:13–27.
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi:10.1126/science.aaa8172.
- Henick BS, Herbst RS, Goldberg SB. The PD-1 pathway as a therapeutic target to overcome immune escape mechanisms in cancer. Expert Opin Ther Targets. 2014;18(12):1407–1420. doi:10.1517/14728222.2014.955794.
- Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32(3–4):643–671. doi:10.1007/s10555-013-9444-6.
- Obrink B. CEA adhesion molecules: multifunctional proteins with signal-regulatory properties. Curr Opin Cell Biol. 1997;9(5):616–626. doi:10.1016/S0955-0674(97)80114-7.
- Kuespert K, Pils S, Hauck CR. CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol. 2006;18(5):565–571. doi:10.1016/j.ceb.2006.08.008.
- Gur C, Maalouf N, Gerhard M, Singer BB, Emgard J, Temper V, Neuman T, Mandelboim O, Bachrach G. The Helicobacter pylori HopQ outermembrane protein inhibits immune cell activities. Oncoimmunology. 2019;8(4):e1553487. doi:10.1080/2162402X.2018.1553487.
- Gur C, Maalouf N, Shhadeh A, Berhani O, Singer BB, Bachrach G, Mandelboim O. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology. 2019;8(6):e1581531. doi:10.1080/2162402X.2019.1581531.
- Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol. 2006;6(6):433–446. doi:10.1038/nri1864.
- Kammerer R, Hahn S, Singer BB, Luo JS, Von Kleist S, Von Kleist S. Biliary glycoprotein (CD66a), a cell adhesion molecule of the immunoglobulin superfamily, on human lymphocytes: structure, expression and involvement in T cell activation. Eur J Immunol. 1998;28(11):3664–3674. doi:10.1002/(SICI)1521-4141(199811)28:11<3664::AID-IMMU3664>3.0.CO;2-D.
- Singer BB, Scheffrahn I, Heymann R, Sigmundsson K, Kammerer R, Obrink B. Carcinoembryonic antigen-related cell adhesion molecule 1 expression and signaling in human, mouse, and rat leukocytes: evidence for replacement of the short cytoplasmic domain isoform by glycosylphosphatidylinositol-linked proteins in human leukocytes. J Immunol. 2002;168(10):5139–5146. doi:10.4049/jimmunol.168.10.5139.
- Witzens-Harig M, Hose D, Junger S, Pfirschke C, Khandelwal N, Umansky L, Seckinger A, Conrad H, Brackertz B, Rème T, et al. Tumor cells in multiple myeloma patients inhibit myeloma-reactive T cells through carcinoembryonic antigen-related cell adhesion molecule-6. Blood. 2013;121(22):4493–4503. doi:10.1182/blood-2012-05-429415.
- Blumenthal RD, Leon E, Hansen HJ, Goldenberg DM. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7(1):2. doi:10.1186/1471-2407-7-2.
- Deng X, Liu P, Zhao Y, Wang Q Expression profiling of CEACAM6 associated with the tumorigenesis and progression in gastric adenocarcinoma. Genet Mol Res. 2014;13(3):7686–7697. doi:10.4238/2014.September.26.6.
- Duxbury MS, Matros E, Clancy T, Bailey G, Doff M, Zinner MJ, Ashley SW, Maitra A, Redston M, Whang EE, et al. CEACAM6 is a novel biomarker in pancreatic adenocarcinoma and PanIN lesions. Ann Surg. 2005;241(3):491–496. doi:10.1097/01.sla.0000154455.86404.e9.
- Jantscheff P, Terracciano L, Lowy A, Glatz-Krieger K, Grunert F, Micheel B, Brümmer J, Laffer U, Metzger U, Herrmann R, et al. Expression of CEACAM6 in resectable colorectal cancer: a factor of independent prognostic signi ficance. J Clin Oncol. 2003;21(19):3638–3646. doi:10.1200/JCO.2003.55.135.
- Strickland LA, Ross J, Williams S, Ross S, Romero M, Spencer S, Erickson R, Sutcliffe J, Verbeke C, Polakis P, et al. Preclinical evaluation of carcinoembryonic cell adhesion molecule (CEACAM) 6 as potential therapy target for pancreatic adenocarcinoma. J Pathol. 2009;218(3):380–390. doi:10.1002/path.2545.
- Kim KS, Kim JT, Lee SJ, Kang MA, Choe IS, Kang YH, Kim S-Y, Yeom YI, Lee Y-H, Kim JH, et al. Overexpression and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 6 in colorectal cancer. Clin Chim Acta. 2013;415:12–19. doi:10.1016/j.cca.2012.09.003.
- Kobayashi M, Miki Y, Ebina M, Abe K, Mori K, Narumi S, Suzuki T, Sato I, Maemondo M, Endo C, et al. Carcinoembryonic antigen-related cell adhesion molecules as surrogate markers for EGFR inhibitor sensitivity in human lung adenocarcinoma. Br J Cancer. 2012;107(10):1745–1753. doi:10.1038/bjc.2012.422.
- Zang M, Hu L, Cao S, Fan Z, Pang L, Li J, Su L, Li C, Liu W, Gu Q, et al. Dual role of carcinoembryonic antigen-related cell adhesion molecule 6 expression in predicting the overall survival of gastric cancer patients. Sci Rep. 2017;7(1):10773. doi:10.1038/s41598-017-11482-9.
- Zang M, Zhang B, Zhang Y, Li J, Su L, Zhu Z, Gu Q, Liu B, Yan M. CEACAM6 promotes gastric cancer invasion and metastasis by inducing epithelial-mesenchymal transition via PI3K/AKT signaling pathway. PLoS One. 2014;9(11):e112908. doi:10.1371/journal.pone.0112908.
- Burtin P, Quan PC, Sabine MC Nonspecific cross reacting antigen as a marker for human polymorphs, macrophages and monocytes. Nature. 1975;255(5511):714–716. doi:10.1038/255714a0.
- Liu Y, Xia T, Jin C, Gu D, Yu J, Shi W, Zhang KE, Zhang L, Ye J, Li L, et al. FOXP3 and CEACAM6 expression and T cell infiltration in the occurrence and development of colon cancer. Oncol Lett. 2016;11(6):3693–3701. doi:10.3892/ol.2016.4439.
- Kuroki M, Matsuo Y, Kinugasa T, Matsuoka Y Three different NCA species, CGM6/CD67, NCA-95, and NCA-90, are comprised in the major 90 to 100-kDa band of granulocyte NCA detectable upon SDS-polyacrylamide gel electrophoresis. Biochem Biophys Res Commun. 1992;182(2):501–506. doi:10.1016/0006-291X(92)91760-N.
- Kolla V, Gonzales LW, Bailey NA, Wang P, Angampalli S, Godinez MH, Madesh M, Ballard PL Carcinoembryonic cell adhesion molecule 6 in human lung: regulated expression of a multifunctional type II cell protein. Am J Physiol Lung Cell Mol Physiol. 2009;296(6):L1019–30. doi:10.1152/ajplung.90596.2008.
- Chan CH, Stanners CP Novel mouse model for carcinoembryonic antigen-based therapy. Mol Ther. 2004;9(6):775–785. doi:10.1016/j.ymthe.2004.03.009.
- Scholzel S, Zimmermann W, Schwarzkopf G, Grunert F, Rogaczewski B, Thompson J Carcinoembryonic antigen family members CEACAM6 and CEACAM7 are differentially expressed in normal tissues and oppositely deregulated in hyperplastic colorectal polyps and early adenomas. Am J Pathol. 2000;156(2):595–605. doi:10.1016/S0002-9440(10)64764-5.
- Kammerer R, Zimmermann W. Coevolution of activating and inhibitory receptors within mammalian carcinoembryonic antigen families. BMC Biol. 2010;8(1):12. doi:10.1186/1741-7007-8-12.
- Pavlopoulou A, Scorilas A A comprehensive phylogenetic and structural analysis of the carcinoembryonic antigen (CEA) gene family. Genome Biol Evol. 2014;6(6):1314–1326. doi:10.1093/gbe/evu103.
- Daniel S, Nagel G, Johnson JP, Lobo FM, Hirn M, Jantscheff P, Kuroki, M, von Kleist, S, Grunert, F. Determination of the specificities of monoclonal antibodies recognizing members of the CEA family using a panel of transfectants. Int J Cancer. 1993;55(2):303–310. doi:10.1002/ijc.2910550222.
- Jantscheff P, Nagel G, Thompson J, Kleist SV, Embleton MJ, Price MR, Grunert, F.A CD66a-specific, activation-dependent epitope detected by recombinant human single chain fragments (scFvs) on CHO transfectants and activated granulocytes. J Leukoc Biol. 1996;59(6):891–901. doi:10.1002/jlb.59.6.891.
- Du X, Luka J, Stafford LJ, Semenuk M, Wang X-P, Kantor J, Bristol , AJ.Colon and pancreas cancer specific antigens and antibodies. Patent US20130189268. 2013.
- Fantini M, David JM, Saric O, Dubeykovskiy A, Cui Y, Mavroukakis SA, Bristol A, Annunziata CM, Tsang KY, Arlen PM, et al. Preclinical characterization of a novel monoclonal antibody NEO-201 for the treatment of human carcinomas. Front Immunol. 2018;8:1899. doi:10.3389/fimmu.2017.01899.
- Zeligs K, Arlen PM, Tsang K, Hernandez L, Fantini M, Annunziata CM. Preclinical characterization of a novel monoclonal antibody targeting a neo-antigen expressed in ovarian and GI malignancies. Cancer Res. 2017; 77:3025.
- Markel G inventor. Antibodies to carcinoembryonic antigen-related cell adhesion molecule (CEACAM). Patent WO2013054320. 2013.
- Brackertz B, Conrad H, Daniel J, Kast B, Kronig H, Busch DH, Adamski, J, Peschel, C, Bernhard, H.FLT3-regulated antigens as targets for leukemia-reactive cytotoxic T lymphocytes. Blood Cancer J. 2011;1(3):e11. doi:10.1038/bcj.2011.12.
- Moosmann A. Gezielte Reaktivierung spezifischer zytotoxischer T-Zellen mit Epstein-Barr-Virus-Vektoren. Germany: Ludwig-Maximilians-University Munich; 2002.
- Kuroki M, Matsuo Y, Kinugasa T, Matsuoka Y Augmented expression and release of nonspecific cross-reacting antigens (NCAs), members of the CEA family, by human neutrophils during cell activation. J Leukoc Biol. 1992;52(5):551–557. doi:10.1002/jlb.52.5.551.
- Poschke I, Faryna M, Bergmann F, Flossdorf M, Lauenstein C, Hermes J, Hinz, U, Hank, T, Ehrenberg, R, Volkmar, M, et al. Identification of a tumor-reactive T-cell repertoire in the immune infiltrate of patients with resectable pancreatic ductal adenocarcinoma. Oncoimmunology. 2016;5(12):e1240859. doi:10.1080/2162402X.2016.1240859.
- Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, Kang, B, Liu, Z, Jin, L, Xing, R, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. 2018;24(7):978–985. doi:10.1038/s41591-018-0045-3.
- Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, Leeson R, Kanodia A, Mei S, Lin J-R, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175(4):984–97 e24. doi:10.1016/j.cell.2018.09.006.
- Chen Z, Chen L, Qiao SW, Nagaishi T, Blumberg RS Carcinoembryonic antigen-related cell adhesion molecule 1 inhibits proximal TCR signaling by targeting ZAP-70. J Immunol. 2008;180(9):6085–6093. doi:10.4049/jimmunol.180.9.6085.
- Reissfelder C, Stamova S, Gossmann C, Braun M, Bonertz A, Walliczek U, Grimm M, Rahbari NN, Koch M, Saadati M, et al. Tumor-specific cytotoxic T lymphocyte activity determines colorectal cancer patient prognosis. J Clin Invest. 2015;125(2):739–751. doi:10.1172/JCI74894.
- Slevogt H, Zabel S, Opitz B, Hocke A, Eitel J, N’Guessan PD, Lucka L, Riesbeck K, Zimmermann W, Zweigner J, et al. CEACAM1 inhibits Toll-like receptor 2-triggered antibacterial responses of human pulmonary epithelial cells. Nat Immunol. 2008;9(11):1270–1278. doi:10.1038/ni.1661.
- Khairnar V, Duhan V, Maney SK, Honke N, Shaabani N, Pandyra AA, Seifert M, Pozdeev V, Xu HC, Sharma P, et al. CEACAM1 induces B-cell survival and is essential for protective antiviral antibody production. Nat Commun. 2015;6:6217. doi:10.1038/ncomms7217.
- Xydia M, Rahbari R, Ruggiero E, Macaulay I, Tarabichi M, Lohmayer R, Wilkening S, Michels T, Brown D, Vanuytven S, et al. Common clonal origin of conventional T cells and induced regulatory T cells in breast cancer patients. Nat Commun. 2021;12(1):1119. doi:10.1038/s41467-021-21297-y.
- Ramani SR, Tom I, Lewin-Koh N, Wranik B, Depalatis L, Zhang J, Eaton D, Gonzalez LC A secreted protein microarray platform for extracellular protein interaction discovery. Anal Biochem. 2012;420(2):127–138. doi:10.1016/j.ab.2011.09.017.
- Nagaishi T, Pao L, Lin SH, Iijima H, Kaser A, Qiao SW, Chen Z, Glickman J, Najjar SM, Nakajima A, et al. SHP1 phosphatase-dependent T cell inhibition by CEACAM1 adhesion molecule isoforms. Immunity. 2006;25(5):769–781. doi:10.1016/j.immuni.2006.08.026.
- Shapira R, Weber JS, Geva R, Sznol M, Kluger HM, Wong DJL, Liang BC A phase I, open-label, multicenter, single-dose escalation and multi-dose study of a monoclonal antibody targeting CEACAM1 in subjects with selected advanced or recurrent malignancies. Journal of Clinical Oncology. 2020;38(15suppl). doi:10.1200/JCO.2020.38.15_suppl.3094.
- De La Rochere P, Guil-Luna S, Decaudin D, Azar G, Sidhu SS, Piaggio E Humanized mice for the study of immuno-oncology. Trends Immunol. 2018;39(9):748–763. doi:10.1016/j.it.2018.07.001.
- Stripecke R, Munz C, Schuringa JJ, Bissig KD, Soper B, Meeham T, Yao, L-C, Di Santo, JP, Brehm, M, Rodriguez, E, et al. Innovations, challenges, and minimal information for standardization of humanized mice. EMBO Mol Med. 2020;12(7):e8662. doi:10.15252/emmm.201708662.

