Abstract
Pharmacometabonomics describes the use of metabolic profiling of biofluids, tissues and tissue extracts to predict, prior to dosing, the beneficial and adverse effects of an intervention such as drug administration. The approach not only is analogous to pharmacogenomics but also is sensitive to environmental factors such as the gut microbiome. Recent applications of pharmacometabonomics are presented and the extension to the use of longitudinal sampling is introduced. Clinical and other human applications of pharmacometabonomics are highlighted and possible future clinical applications of pharmacometabonomics and longitudinal pharmacometabonomics are discussed. These include clinical trials of new drugs either at the first-into-man stage or later in Phase II and III trials, and assessment of individual patients or groups of patients for particular therapies (personalised and stratified medicine approaches). Since metabonomics approaches are sensitive to both the host genome effects and the gut microbiome, pharmacometabonomics has particular utility for studying the host–microbiome interactions and for assessing new therapies that target the gut bacteria. Since the microbiome also has implications for nutrition and drug pharmacokinetics, such metabolic profiling approaches are likely to of use in such studies. It is anticipated that as metabonomics analytical and statistical technologies continue to develop, more applications will be realised and these should find use in real clinical situations, even monitoring patients in real time.
1. Pharmacometabonomics
In the 21st century health care, one major thrust will be to provide more effective therapies to individual patients in order to minimise adverse effects and to maximise efficacy for a given individual. Although it is unlikely that a drug that is suitable only for a few individuals (truly personalised medicine) will be developed, it is now clear that stratified medicine, that is, tailoring a therapy to selected groups of patients based on some prognostic measure of how they will respond, is a major goal today. This requires systems to predict i) how a given patient will metabolise any administered drugs, ii) whether there will be any adverse side effects and iii) whether the drugs will be efficacious Citation[1]. This approach extends beyond drug therapy and is also relevant to surgical and other interventions such as radiotherapy in cancer treatment, to nutritional and nutraceutical studies, or sports training regimes, for example. To achieve such an approach requires a system of patient evaluation that would tell clinicians the correct drug, dose or intervention for any individual before the start of therapy.
Most personalised approaches have so far been mainly based on measuring differences in genes related to susceptibility to cancer Citation[2] or to polymorphisms in drug-metabolising enzymes such as cytochrome P450 isoenzymes and N-acetyl transferases. There are numerous examples of adverse drug reactions linked to specific enzymatic deficiencies or mutations and thus personalised therapy medicine based on genetic knowledge has been the first line of attack, a paradigm known as pharmacogenomics Citation[3].
However, such methods are limited because most major diseases are a complex interplay between genetic and environmental influences. At an individual level, such external environmental influences can affect drug metabolism, toxicity and treatment efficacy.
In an alternative, but complementary approach at the biological level of small-molecule metabolites, pharmacometabonomics has now been used to understand and predict the interventional outcome of drugs (such as toxicity and xenobiotic metabolism in animal model systems) based on mathematical models derived from pre-dose, biofluid metabolite profiles Citation[4].
Most metabonomics studies have employed nuclear magnetic resonance (NMR) or mass spectrometry (MS), the latter preceded by a liquid chromatography separation. Although the capital cost of such equipment is high, the possible throughput makes the cost per sample comparable with conventional assays.
Initially, the effects of three structurally diverse hepatotoxins in rats (galactosamine, allyl alcohol and paracetamol), which act via different mechanisms were studied, and it was found that pre-dose urinary profiles carried information about the degree of post-dosing toxicity () and, in the case of paracetamol, information about the inter-animal variation in drug metabolism as well Citation[4]. Subsequently, the first demonstration of pharmacometabonomics in humans showed a clear connection between an individual's pre-dose urinary metabolic phenotype and the metabolic fate of a standard dose of the widely used analgesic acetaminophen (paracetamol) Citation[5]. The metabolic fate of the drug was obtained from post-dose urine samples, determining the proportions of the various drug metabolites excreted by each subject, this being known to show considerable intersubject variation. This is important because how a particular drug is metabolised and excreted by each individual can have a major influence on its safety and efficacy. Also Winnike et al. showed that well-known enzyme markers of toxicity were predictable from early post-dose profiles Citation[6]. In addition, it has been shown that pharmacokinetics of tacrolimus can be predicted from pre-dose metabolic analyses Citation[7]. One area where such an approach might be useful but in which pharmacometabonomics has not yet found application is attempting to predict the combined effects of multiple drugs dosed simultaneously, that is, polypharmacy, which is a common regime, especially in older people. This approach could also have application in other mixed-agent therapeutic approaches such as traditional Chinese medicine, herbal mixtures and multifaceted dietary interventions.
Figure 1. The first exemplification of the pharmacometabonomics concept based on pre-dose prediction of toxicity in an animal model. (Top) Typical 1H NMR spectrum of urine before dosing with the toxin galactosamine. (Bottom) A principal components scores plot where each point represents a single rat urine taken before dosing. Red indicates rats that subsequently showed high toxicity and green points are for rats that showed no toxicity. Although there is a partial overlap between the two groups of animals, a clear distinction is observed Citation[4].
![Figure 1. The first exemplification of the pharmacometabonomics concept based on pre-dose prediction of toxicity in an animal model. (Top) Typical 1H NMR spectrum of urine before dosing with the toxin galactosamine. (Bottom) A principal components scores plot where each point represents a single rat urine taken before dosing. Red indicates rats that subsequently showed high toxicity and green points are for rats that showed no toxicity. Although there is a partial overlap between the two groups of animals, a clear distinction is observed Citation[4].](/cms/asset/dbb2289e-28c1-460c-b310-45be36cfa9d8/iemt_a_11103357_f0001_b.jpg)
These pharmacometabonomics studies imply the possibility of applying this approach prospectively to screening humans in populations to predict disease onset, but this is still a long way off and, as with human genomic studies, there also significant ethical issues involved with such screening procedures in man.
2. Longitudinal pharmacometabonomics
However, it is practical to screen and follow longitudinally patients who present for a particular type of therapy such as cancer chemotherapy or radiotherapy. In a recent Lancet article, Kinross et al. proposed such a clinical phenotyping process for monitoring patient journeys before, during and after surgery Citation[8]. Thus we can distinguish between pharmacometabonomics, which is the use of steady-state, pre-dose metabolic profiling to predict drug metabolism, adverse effects and efficacy, and longitudinal pharmacometabonomics, which involves the measurement of a person's metabolic profiles prior to, during and after some intervention, in order to stratify the patient in terms of prediction of response to future treatments (e.g., prognosis of the efficacy or adverse side effects of chemotherapy) or the likelihood of any outcome (e.g., need for intensive care facilities). The differences in metabolic profiles for a given patient at different times can be considered as a metabolic trajectory. We anticipate that the differences in metabolic trajectories between patients will lead to better stratification of treatments. This extension to pharmacometabonomics holds out the promise of significant benefits for patients in terms of efficacy and safety and also for health service providers in terms of cost and efficiency Citation[8]. How this could work in practice for surgery is encapsulated in .
Figure 2. A schematic diagram indicating how longitudinal pharmacometabonomics could be used to monitor and stratify patients who are undergoing surgery. Samples can be taken at all stages of patient treatment and used to provide metabolic trajectory information, i.e., the changes in metabolic profiles over time can be used to predict patient outcome and to guide the physician and surgeon. The patient's metabolic trajectory will reflect the combined effects of disease progression and clinical therapy. Both short-term stratification and risk assessment are possible and also, in principle, long-term patient management. Finally, samples can be biobanked so as to build up a large collection suitable for epidemiological study.
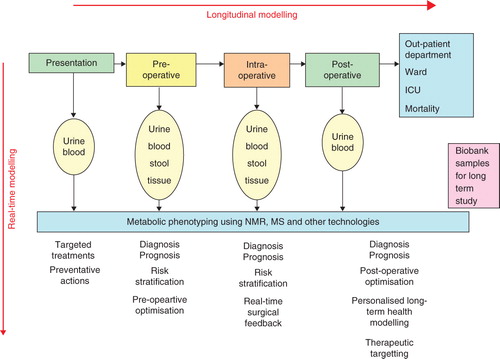
The minimum number of samples required to build robust predictive models varies according to the application. In the case of human population metabonomic profiling, it has been shown that a few hundred individuals are sufficient. For the human pharmacometabonomics study of paracetamol metabolism, 99 normal, healthy, non-smoking male subjects were used successfully Citation[5]. For more diverse populations such as hospital inpatients, larger sample numbers, formulated using appropriate statistical power calculations, will be required so that all of the normal and abnormal sources of human metabolic variation are well sampled Citation[9]. In clinical longitudinal pharmacometabonomics, it might not be necessary to provide a high level of predictive precision, and, for example, it might be sufficient to give a simple output such as a low, medium or high risk of an adverse reaction with suitable ranges. In addition, because we are dealing with metabolic trajectories as opposed to static metabolic profiles, a much greater amount of information and higher quality of information is available for each patient.
3. Expert opinion
While it is recognised that there are no current real-time clinical applications, it can be envisaged that one likely near-term implementation of pharmacometabonomics could be in the realm of pharmaceutical clinical trials, either at the first-in-human healthy volunteer stage or later in patients in Phase II and III trials. Pre-dose metabolic models could be built and then related to quantitative metabolic fates of compounds, any observed adverse reactions or intersubject variation in efficacy.
Moreover, the effects of dietary modulation, pre-biotic and probiotic treatments and other lifestyle changes could also ultimately be evaluated in this way Citation[10]. This is important because ‘personalised health care’ means different things to different people and in some cases lifestyle management not drug therapy, which may be effective for disease prevention, can be a better outcome than the need to find a therapeutic solution after a disease has arisen.
If the pharmacometabonomic predictors for a particular drug action involve host–microbiome co-metabolites, such as was found in the case of the human metabolism of paracetamol, then this clearly promotes the idea of the gut microbiome being a new target for pharmaceutical interventions Citation[11]. Indeed, the targeting of a bacterial enzyme has recently proved successful for alleviating toxicity after cancer chemotherapy Citation[12]. Other possibilities also include the manipulation of the gut microflora to affect the pharmacokinetics and metabolism of drugs in a beneficial manner. For example, many drug hydroxyl groups are conjugated via sulfonation reactions, to form sulfate conjugates, to aid drug elimination. Drug sulfonation in the human body will be affected by competition with 4-cresol, a gut microfloral metabolite, which is subject to sulfonation itself and excretion in the urine as 4-cresol sulfate at millimolar concentrations Citation[13]. Since drug conjugation by sulfonation is a common mechanism, the effect of microbial 4-cresol excretion on drug metabolism should be observable in high-dose drugs other than paracetamol, which undergo excretion after sulfation. Thus, we expect 4-cresol sulfate to be a biomarker for the prediction of the metabolism in such drugs. This recent finding also has implications for diseases such as autism where abnormal sulfur metabolism has been implicated. Since sulfur chemistry is integral to glutathione biosynthesis, and this is highly important in maintaining the redox balance, any of the many diseases where this is abnormal could be sensitive to such effects and pharmacometabonomics might have an especially clear role there.
Thus, one main use of a pharmacometabonomic approach would be to act as a screening tool for selecting individuals according to their suitability for treatment with particular drugs, drug classes or drug doses, or other types of therapy. In particular, pharmacometabonomics could potentially provide a gateway to stratified medicine. One very recent study has successfully shown that basal metabolite levels can be prognostic for a future condition Citation[14]. In this case, it was possible to predict future toxicity based on the NCI criteria in patients undergoing chemotherapy with capecitabine for colorectal cancer based on certain lipid-related serum metabolite levels. In addition, use of the approach for investigation of cytochrome P450 allele variation is an interesting option given the large effects on drug metabolism and hence on effectiveness of personalised therapy.
Such applications should be aided by recent developments in data analysis and biomarker identification Citation[15] and the use of analytical techniques other than the main LC–MS and NMR approaches to sample more of the metabolic hyperspace, particularly lower concentration substances. Metabolite profiling of fluids other than urine, such as blood, and particularly, faecal extracts, is expected to provide additional information and such results should be used in an integrated fashion.
In practice, the success of the pharmacometabonomic approach would be expected to vary from drug to drug and would depend on the nature of the challenge posed by each drug, and also on the response of interest. However, certain classes of drugs may be subject to similar predictive models. In principle, by using this methodology, adverse drug reactions could potentially be avoided and drugs and dose levels could be targeted more effectively according to the metabolic and other characteristics of each individual.
It is recognised that optimal stratification of drug treatments will probably involve a variety of complementary response-prediction approaches, including pharmacometabonomics, pharmacogenomics and metagenomics of the gut microbiota. Pharmacogenomics has already found application in drug and theranostic development and for identifying individuals for treatment. Pharmacometabonomics has some way to go to achieve this track record, but it has an important theoretical advantage over pharmacogenomics in that it can potentially take account of both genomic and environmental factors affecting drug-induced responses. Furthermore, although pharmacometabonomics would normally relate to predicting drug- or xenobiotic-induced responses, we envisage that similar methodology could also be applied to predicting individual responses to broader medical, dietary, microbiological or physiological challenges.
The exciting new concept of longitudinal pharmacometabonomics involves the tracking of a patient's metabolic profile over time. Although the approach has not yet been validated in the clinic, the patient's metabolic trajectory will reflect both disease progression and the results of clinical therapy and, we believe, will allow patient stratification in terms of the prediction of outcome and the choice of optimal treatment.
Declaration of interest
JK Nicholson and JR Everett are supported by their institutions. All authors are named inventors on a granted patent describing pharmacometabonomics and metabolic phenotyping (No. EP1540560).
Bibliography
- Scheiber J. How can we enable drug discovery informatics for personalized healthcare. Expert Opin Drug Discov 2011;6:219-24
- Yong WP, Innocenti F Ratain MJ. The role of pharmacogenetics in cancer therapeutics. Br J Clin Pharmacol 2006;62:35-46
- Watson RG, McLeod HL. Pharmacogenomic contribution to drug response. Cancer J 2011;17:80-8
- Clayton TA, Lindon JC, Cloarec O, Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature 2006;440:1073-7
- Clayton TA, Baker D, Lindon JC, Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci USA 2009;106:14728-33
- Winnike JH, Wright FA, Macdonald JM, Use of pharmaco-metabonomics for early prediction of acetaminophen-induced hepatotoxicity in humans. Clin Pharm 2010;88:45-51
- Phapale PB, Kim SD, Lee HW, An integrated approach for identifying a metabolic phenotype predictive of individualized pharmacokinetics of tacrolimus. Clin Pharmacol Ther 2010;87:426-36
- Kinross JM, Holmes E, Darzi AW, Metabolic phenotyping in monitoring of surgical patients. Lancet 2011;377:1817-19
- Nicholson G, Rantalainen M, Maher AD, Human metabolic profiles are stably controlled by genetic and environmental variation. Mol Syst Biol 2011;7:525
- Rezzi S, Ramadan Z, Martin F-PJ, Human metabolic phenotypes link directly to specific dietary preferences in healthy individuals. J Proteome Res 2007;6:4469-77
- Jia W, Li H, Zhao L, Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov 2008;7:123-9
- Wallace BD, Wang H, Lane KT, Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010;330:831-5
- Yap IKS, Angley M, Veselkov KA, Urinary metabolic phenotyping differentiates children with autism from their unaffected siblings and age-matched controls. J Proteome Res 2010;9:2996-3004
- Backshall A, Sharma R, Clarke SJ, Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin Cancer Res 2011;17:3019-28
- Lindon JC, Nicholson JK. Spectroscopic and statistical techniques for information recovery in metabonomics and metabolomics. Ann Rev Anal Chem 2008;1:45-69
