Abstract
Introduction
Ultrasensitive bacterial detection methods are crucial to ensuring accurate diagnosis and effective clinical monitoring, given the significant threat bacterial infections pose to human health. The aim of this study is to develop a biosensor with capabilities for broad-spectrum bacterial detection, rapid processing, and cost-effectiveness.
Methods
A magnetically-assisted SERS biosensor was designed, employing wheat germ agglutinin (WGA) for broad-spectrum recognition and antibodies for specific capture. Gold nanostars (AuNSs) were sequentially modified with the Raman reporter molecules and WGA, creating a versatile SERS tag with high affinity for a diverse range of bacteria. Staphylococcus aureus (S. aureus) and Pseudomonas aeruginosa (P. aeruginosa) antibody-modified Fe3O4 magnetic gold nanoparticles (MGNPs) served as the capture probes. Target bacteria were captured by MGNPs and combined with SERS tags, forming a “sandwich” composite structure for bacterial detection.
Results
AuNSs, with a core size of 65 nm, exhibited excellent storage stability (RSD=5.6%) and demonstrated superior SERS enhancement compared to colloidal gold nanoparticles. Efficient binding of S. aureus and P. aeruginosa to MGNPs resulted in capture efficiencies of 89.13% and 85.31%, respectively. Under optimized conditions, the developed assay achieved a limit of detection (LOD) of 7 CFU/mL for S. aureus and 5 CFU/mL for P. aeruginosa. The bacterial concentration (10–106 CFU/mL) showed a strong linear correlation with the SERS intensity at 1331 cm−1. Additionally, high recoveries (84.8% - 118.0%) and low RSD (6.21% - 11.42%) were observed in spiked human urine samples.
Conclusion
This study introduces a simple and innovative magnetically-assisted SERS biosensor for the sensitive and quantitative detection of S. aureus or P. aeruginosa, utilizing WGA and antibodies. The developed biosensor enhances the capabilities of the “sandwich” type SERS biosensor, offering a novel and effective platform for accurate and timely clinical diagnosis of bacterial infections.
Introduction
Bacterial infections continue to pose a significant public health concern, affecting an estimated 258 million people worldwide and resulting in nearly 280,000 deaths annually.Citation1,Citation2 Current methods, including plate culture,Citation3 polymerase chain reaction (PCR),Citation4 and enzyme-linked immunosorbent assays (ELISA),Citation5 are commonly employed for pathogen detection. These methods for bacterial detection typically take at least 48 hours to complete, involving processes such as isolation, cultivation, and identification of the bacteria. This process can be time-consuming, especially for slowly growing bacteria. Moreover, the steps involving separation and enrichment are time-consuming, potentially causing delays in achieving final and accurate bacterial detection.Citation6 Delays in such situations are undesirable, particularly in emergency conditions, as they pose a serious risk to patients’ lives and health.
Several promising biosensors based on fluorescence, chemiluminescence, electrochemistry, and surface plasmon resonance have been developed to address challenges in clinical microbial testing.Citation7,Citation8 Among these methods, label-based surface-enhanced Raman scattering (SERS) stands out as a potent analytical technique extensively applied in bacterial detection.Citation9–11 SERS tags, at the core of SERS biosensors, are nanoprobes with SERS activity, capable of recognizing the target and generating a significant and specific Raman signal.Citation12 Typical SERS tags include biorecognition elements for identifying target bacteria, precious metal nanomaterials, and specific Raman reporter molecules.Citation13 For instance, Zhu et al modified aptamer and 4-aminothiophenol (4-ATP) on gated aminated mesoporous silica nanoparticles (MSNs) as a SERS substrate for detecting Staphylococcus aureus (S. aureus).Citation14 Zhuang et al designed ultra-sensitive SERS tags using gold nanostars (AuNSs) and 4-mercaptobenzoic acid (4-MBA) for detecting Salmonella typhimurium (S. typhimurium) in milk and meat samples.Citation15
Currently, the development of such methods faces a critical and complex challenge concerning the preparation of SERS tags, requiring good stability, substantial Raman signal enhancement, and efficient binding to target bacteria. However, conventional colloidal gold and colloidal silver nanoparticles exhibit limited signal amplification. Additionally, the production of multifunctional nanoparticles is both time-consuming and challenging.Citation16 In the construction of dual-recognition SERS biosensors, it is often necessary to employ a pair of biorecognition molecules for identifying the target bacteria. Antibodies,Citation17 aptamers,Citation18 antibioticsCitation19 (eg, vancomycin), and antimicrobial peptidesCitation20 are among the most widely used biorecognition agents. Antibodies, especially monoclonal antibodies, are highly homogeneous and recognize only a specific antigenic epitope. The high affinity allows antibodies to have higher detection sensitivity and binding ability to low concentrations of target molecules, making them one of the most commonly used types of antibodies in current in vitro diagnostic studies. However, antibodies are produced through animal immunization and can incur high costs when employed in assay labels requiring a large number of recognition elements.Citation21 Furthermore, aptamers require additional time and specific ions to unfold their spatial structure, posing challenges in screening for aptamers with high binding affinity to the target bacteria.Citation22 Consequently, the scarcity of dependable bacterial identification elements restricts the widespread application of label-based SERS detection.
In recent research, lectins have emerged as promising agents for pathogen capture, given their favorable characteristics such as low cost, high stability, and widespread availability.Citation23 Wheat germ agglutinin (WGA), one of the most researched and least immunogenic lectins, binds explicitly to N-acetyl-D-glucosamine and N-acetyl-D-glucosamine derivatives on bacterial surfaces.Citation24,Citation25 Considering the prevalence of polysaccharides on bacterial surfaces, WGA exhibits immense potential for diverse applications in bacterial detection.Citation26 For example, Yang et al successfully and affordably identified S. aureus in blood samples by modifying WGA on magnetic gold nanoparticles (MGNPs) and combining it with a colorimetric method.Citation27 Tu et al employed WGA-modified magnetic quantum dots as a versatile assay nanoprobe to create a highly sensitive, multiplexed lateral flow assay for detecting Pseudomonas aeruginosa (P. aeruginosa) and S. typhimurium in complex samples.Citation28
To enhance the detection sensitivity of SERS biosensors, a novel dual-recognition SERS biosensor was designed by incorporating WGA with high-performance AuNSs. Model bacteria, including Gram-positive S. aureus and Gram-negative P. aeruginosa, were selected to assess the universal detection capability of the platform. The developed approach involves three key steps. Firstly, target bacteria were captured by MGNPs modified with antibodies. Subsequently, WGA-SERS tags were introduced into the solution to form a sandwich structure (MGNPs/bacteria/SERS tags). The resulting SERS signal from this sandwich structure was promptly analyzed using Raman spectroscopy. Furthermore, the platform demonstrates versatility in detecting various pathogenic bacteria, allowing the easy replacement of specific antibodies tailored to different bacterial strains. We anticipate that this method will provide valuable insights into universal testing for the diagnosis of clinical bacterial infections.
Materials and Methods
Materials
5.5’-dithiobis-(2-nitrobenzoic acid) (DTNB), Bovine serum albumin (BSA), Polyethylenimine (PEI, MW ~25 kDa), Polyvinylpyrrolidone (PVP, MW 40 K), N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide (EDC), N-hydroxy-sulfosuccinimide (sulfo-NHS), 2-(N-morpholino) ethanesulfonic acid (MES), phosphate buffered saline (PBS), Tween-20, and Wheat germ agglutinin lectin from Triticum vulgaris (WGA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Chloroauric acid tetrahydrate (HAuCl4·4H2O), sodium borohydride (NaBH4), 1×Phosphate-buffered saline (PBS, pH 7.4), ethanolamine hydroxylamine hydrochloride, hydroquinone, sodium dodecyl sulfate (SDS) and trisodium citrate (TSC) were obtained from Sinopharm Chemical Reagent Co (Shanghai, China). Mouse monoclonal anti-S. aureus antibodies (Catalog no. ab20920) and mouse monoclonal anti-P. aeruginosa antibodies (catalog no. ab68538) were brought from Abcam (Cambridge, United Kingdom).
The UV−vis absorption spectra were recorded using the Shimadzu 2600 spectrometer. Transmission electron microscope (TEM) images of nanocomposites were acquired with a Hitachi H-7650 microscope operating at 80 kV. Scanning electron microscopy (SEM) (JTOL JSM-7001 F) was employed to capture the surface morphology. Raman measurements were performed using a B&W Tek portable Raman spectrometer (i-Raman Plus BWS465-785H) with a 785 nm excitation laser. All Raman spectra were integrated for 10s with 10 mW power.
Acquisition of Bacterial Samples
The standard bacterial strains used in this study, including S. aureus ATCC 25923, P. aeruginosa ATCC 27853, and Escherichia coli (E. coli) ATCC 43888, were purchased from Solarbio Life Science (Beijing, China). Listeria monocytogenes (L. monocytogenes), Klebsiella pneumoniae (K. pneumoniae), and Acinetobacter baumannii (A. baumannii) were provided by the Xiao team from the Beijing Institute of Radiation Medicine.Citation29 Bacterial concentrations were verified using the classical plate count method.Citation30,Citation31 In brief, S. aureus and P. aeruginosa were incubated on 5% sheep blood agar plates for 12 hours. A few colonies were picked from the plate and dispersed in PBS buffer (pH 7.4, 10 mM) to prepare a bacterial suspension. The initial bacterial solution was diluted in sterile water 1×105 to 106 times, and 100 μL of this dilution was coated on the blood agar plate at 37 °C. Colony-forming units (CFU) on the plate were counted after 12 hours of incubation, and the final bacterial solution was further diluted to obtain various experimental concentrations.
MGNPs Assembly
The synthesis pathway of antibody - modified MGNPs is illustrated in . Monodispersed MGNPs were synthesized using the method described in our earlier publication.Citation32–34 Briefly, we opted for Fe3O4 magnetic nanoparticles (MNPs) as the superparamagnetic core to produce MGNPs, leveraging its robust magnetic responsiveness and stability. The synthesis involved the sequential self-assembly of a positively charged polyethyleneimine (PEI) layer and negatively charged 3 nm Au NPs on the MNPs surface to form MNPs-Au seed MNPs. Ultimately, through the in situ reduction of Au3+ on the compact 3 nm Au seed, we achieved the swift formation of uniform Au nanoshells with notable roughness, resulting in multifunctional MGNPs. Second, 20 mM MUA ethanol solution was added under sonication for 20 min for surface carboxylation. Then, the formed MGNPs were resuspended in 1 mL of MES solution (0.1 M, pH 6.0) containing 5 mM EDC under sonication. Third, antibody-modified MGNPs were accomplished through the COOH–NH2 condensation reaction of MUA and antibody. After the reaction for 2 h, 100 μL of BSA solution (10%) was added for an additional 1 h to block superfluous residual carboxyl. Finally, antibody-modified MGNPs were isolated using a magnet, and the isolated particles were rinsed once with PBS before being dispersed in 1 mL of PBS.
Figure 1 Characterization of the synthesized antibody-modified MGNPs. (A) Schematic diagram of the synthesis process of antibody-MGNPs. TEM images of (B) MNPs, (C) MNPs-Au seed, and (D) MGNPs. (E) SEM image of MGNPs. (F) Zeta potentials of the antibody-MGNPs from each stage. (G-J) TEM and SEM images of S. aureus and P. aeruginosa captured by antibody-MGNPs. TEM images of S. aureus (G) and P. aeruginosa (H) with antibody-MGNPs. SEM images of S. aureus (I) and P. aeruginosa (J) with antibody-MGNPs. (K) Capture efficiency of non-modified MGNPs and antibody-MGNPs. ***p < 0.001.

Preparation of WGA-SERS Tags
High-performance AuNSs were synthesized via a seed-mediated growth approach, as illustrated in . First, ~20 nm gold seeds were prepared using a modified Turkevich method. Briefly, 50 μL of HAuCl4 (1%, w/v) and 150 μL of TSC (1%, w/v) were mixed in 5 mL boiling deionized water, stirred for 15 min, and placed at room temperature to cool down. Second, 150 μL of seed solution was mixed with 100 μL of 1% HAuCl4 and 10 mL of deionized water under vigorous stirring. Then, 200 μL of 1% SDS, 50 μL of 1% TSC, and 500 μL of 60 mM hydroquinone were introduced into the solution simultaneously. Finally, the mixture was stirred at room temperature for 20 min to obtain an apparent blue-colored solution.
Figure 2 Characterization and optimization of the synthesized WGA-SERS tags. (A) Schematic diagram of the synthesis process of WGA modified AuNSs@DTNB. TEM images of (B) Au core, and (C) AuNSs. (D) UV-vis spectra of Au core (red) and AuNSs (blue). (E) SERS intensities of AuNSs, AuNSs@DTNB and AuNSs@DTNB@WGA. (F) Zeta potentials in each synthesis stage of WGA-SERS tags. (G) TEM images of AuNSs prepared with different seed amounts. (H) UV-vis spectra of AuNSs prepared with different seed amounts. (I) Raman intensities of the WGA-SERS tags against preservation time.

The WGA-conjugated SERS tags (WGA-SERS tags) were functionalized with AuNSs by using carbodiimide chemistry. In brief, 100 μL of 10−2 M Raman reporters DTNB in ethanol was mixed with 100 mL of AuNSs solution and stirred overnight at room temperature. Then, the prepared AuNSs@DTNB (2 mL) were separated by centrifugation (4500 rpm, 6 min) and then re-suspended in 1 mL of MES buffer (10 mM, pH 5.5). To link the DTNB through amide condensation, for this 5 μL of EDC (10 mM) and 10 μL of NHS (10 mM) were mixed followed by the addition of 2.0 μg WGA for 15 min. At the end of 2 h incubation step, 50 μL of BSA (10%) was added to the mixture and agitated for an additional hour to block unreacted carboxyl groups. The mixture was then isolated by centrifugation at 4500 rpm for 6 min to remove the supernatant. The precipitate was then redispersed in PBS (2 mM, pH 7.4) and labeled as WGA-SERS tags.
Bacteria Detection via SERS Strategy
In a typical experiment, 2 μL of antibody-modified MGNPs (10 mg/mL) and 1 mL of S. aureus or P. aeruginosa solutions within a range of 10–106 CFU/mL were mixed and incubated at 37 °C for 15 min. The non-captured bacteria were removed by magnetic separation and PBS washing twice, and the formed MGNPs/bacteria complexes were suspended in 100 μL of binding buffer (10 mM PBS, 5 mM Ca2+, 5 mM Mg2+). Afterward, the 60 μL of WGA-SERS tags were directly incubated with the above solutions for 15 min on an oscillator at room temperature. Finally, the formed MGNPs/bacteria/SERS tag sandwich complexes were magnetically collected, washed twice with PBS under a magnetic field, and transferred onto a Si chip for subsequent SERS detection using portable Raman spectrometer.
Analysis of Actual Samples
Human urine served as the authentic biological sample. A recovery experiment was performed to enhance the method’s reliability. Samples were promptly collected from three healthy volunteers in our laboratory immediately after voiding. Informed consent was obtained from all volunteers participating in this study, and the study protocol adhered to the principles outlined in the Declaration of Helsinki. The study received approval from the research ethics committee of Guangdong Provincial People’s Hospital (KY-X-2022-003-03).
Various concentrations of S. aureus or P. aeruginosa (5000, 500, 50 CFU/mL) were intentionally introduced into pooled urine samples from healthy volunteers to simulate real-world conditions. Subsequently, 2 μL of antibody-MGNPs were added, and the mixture was oscillated using an oscillator for 15 min. After magnetic separation, 60 μL of WGA-SERS tags were added to form a stable sandwich composite structure. The resulting complexes were analyzed using a portable Raman spectrometer, with each test being repeated three times.
Statistical Analysis
Each experiment was replicated at least three times, and the data were presented as mean ± SD. Statistical analysis employed One-Way ANOVA to assess differences, with significance denoted by p < 0.05.
Results and Discussion
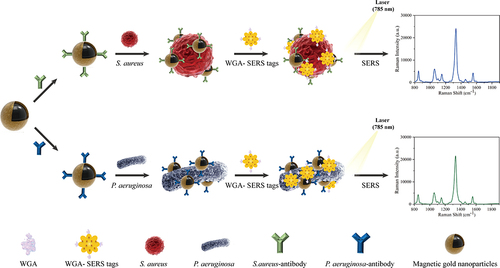
Sensitive detection of S. aureus or P. aeruginosa relies on MGNPs modified with antibodies, serving as capture probes, and AuNSs immobilized with DTNB and WGA, functioning as signal probes. The operational procedure of the dual-recognition SERS biosensor involves three key steps, as illustrated in . First, MGNPs labeled with specific antibodies for either S. aureus or P. aeruginosa are mixed with a sample containing one or more bacterial species. The mixture is then incubated for 15 min, facilitating the specific binding of antibodies to the target bacteria. As specific pathogen recognition probes, MGNPs rely on the specificity of antibodies to concentrate and isolate bacteria using magnetic fields, especially when dealing with complex samples containing multiple bacterial species. Next, the proposed SERS tags are added and incubated for an additional 15 min to form sandwich structures. The AuNSs, modified with WGA as a strong affinity recognition molecule, bind to the peptidoglycan of the bacterial cell wall through strong hydrogen bonding. Finally, the sandwich complexes are collected using an external magnet, rinsed twice with PBST solution, and analyzed for SERS signals using a Raman instrument. Quantitative detection is performed based on the correlation between the characteristic signal intensity of DTNB carried by the sandwich structure and bacterial concentration. Importantly, utilizing high-performance tags and magnetic enrichment leads to a dual enhancement of SERS signals from target bacteria, significantly improving the sensitivity of SERS platforms based on sandwich analysis.
Scheme 1 Schematic illustration of the operating procedure for rapid detection of bacteria by using antibody-modified MGNPs and WGA-SERS tags in combination.

As previously reported, monodisperse MGNPs were synthesized as magnetic separators and SERS substrates using a PEI-mediated seed growth technique. The process of MGNPs synthesis is illustrated in and has been validated through TEM and zeta potential measurements. In , 200 nm superparamagnetic MNPs were initially prepared using the classical solvothermal method. Hydrophilic PEI rapidly self-assembled on the surface of MNPs, forming a charged thin layer that efficiently adsorbed 3 nm gold seeds through electrostatic adsorption (). Subsequently, following the reduction of Au3+ on the surface of the 3 nm gold seeds, a 25 nm layer of gold shells formed on the surface of the MNPs, resulting in the formation of MGNPs (). Additional synthesized MGNPs were visualized in the SEM image (). The MUA molecule, serving as a linker on the surface of MGNPs, binds with antibodies through carbodiimide chemistry. As shown in , the zeta potential of MGNPs modified by MUA decreased from −9.6 mV to −22.9 mV, and dropped back to −15.2 mV after coupling with the antibody, thus indicating the successful binding of the antibody to MGNPs. Subsequent studies explored the effectiveness of this antibody-based magnetic separation system in capturing the target bacteria. After 15 min of incubation with 2 μL of MGNPs and 1000 CFU/mL of S. aureus and P. aeruginosa solutions, the solutions were magnetically separated using an external magnet. The supernatant solution was incubated on blood agar overnight to calculate the capture efficiency, while the binding of bacteria and MGNPs complexes was observed by TEM and SEM. As shown in , MGNPs were tightly bound to S. aureus and P. aeruginosa in the solution. After magnetic separation, colony counting of the supernatant showed capture efficiencies of 89.13% and 85.31% for antibody-modified MGNPs against S. aureus and P. aeruginosa, respectively (), facilitating efficient isolation and enrichment of the target bacteria.
The WGA-SERS tags (AuNSs@DTNB@WGA) were synthesized using a seed-mediated growth strategy, as illustrated in . Initially, Au cores with an average size of 20 nm were synthesized through the sodium citrate reduction method (). Subsequently, TSC and hydroquinone acted as reducing and capping agents on the surface of the Au cores, converting Au3+ to Au0 and enabling the Au cores to grow into star-shaped gold nanostructures with multiple technical angles. The TEM image () revealed nanoscale raised tips on the surface of each AuNS, significantly enhancing the potential “hot spots” in the local field. The formation of star-shaped gold nanostructures caused a shift in the absorption band of the burgundy Au cores in the UV-Vis spectrum from 526 nm to 596 nm, and the solution color turned dark blue (). DTNB molecules covering the surface of AuNSs provided characteristic SERS signals and carried carboxyl groups that could be conjugated to WGA. The SERS spectra of AuNSs, AuNSs@DTNB, and AuNSs@DTNB@WGA (WGA-SERS tags) were measured and analyzed, as shown in . After WGA modification, almost no discernible spectral differences were observed for AuNSs@DTNB, confirming the Raman detection capability of the WGA-modified SERS tags. Successful verification of WGA conjugation with AuNSs was achieved through zeta potential monitoring. The zeta potential of unmodified AuNSs in was −16.5 mV, which decreased further to −23.6 mV after DTNB modification. Subsequently, the zeta potential slightly increased after WGA conjugation, stabilizing at −18.7 mV upon saturation with WGA (exceeding 2 μg), indicating sufficient binding of 2 μg WGA and AuNSs@DTNB.
Controlled volumes of seed solution (50, 100, 150, and 200 μL) were systematically used to synthesize a variety of AuNSs with different sizes. As the volume of the seed solution increased, there were notable alterations in the structure and size of the resulting AuNSs, as illustrated in . The diameter of the AuNSs decreased from 120 nm to 50 nm, accompanied by a gradual blue-shift in the LSPR peak and a transition in solution color from blue to purple (). AuNSs featuring a diameter of 65 nm were selected for subsequent experiments, considering the spatial site resistance effect of SERS tags. Furthermore, the WGA-SERS tags, prepared using these AuNSs, exhibited negligible changes in SERS signals even after storage at 4 °C for 10 days (). The relative standard deviation (RSD) of the Raman signal intensity at 1331 cm−1 was measured at 5.66%, underscoring the commendable storage stability of the freshly prepared WGA-SERS tags.
The practical application of the sensor heavily relies on the effectiveness of the synthesized SERS tags in binding to pathogenic bacteria. In previous studies, we demonstrated that WGA-modified MGNPs exhibited a strong affinity for binding to various pathogenic bacteria, including S. epidermidis, P. aeruginosa, S. aureus, L. mono, E. coli, A. baumannii, S. sonnei, and S. typhi.Citation35,Citation36 In this study, we investigated the binding capability of SERS tags, which were modified with varying levels of WGA, to interact with pathogenic bacteria. The S. aureus model was used as our experimental example. The dynamic process of binding was confirmed through TEM image observation and SERS signal detection using a portable Raman spectrometer ( and ). With an increase in WGA modification from 0.25 μg to 2.5 μg, the number of SERS tags bound to the bacterial surface gradually increased. Notably, at a WGA modification of 2.0 μg, the WGA-SERS tags adhered densely and uniformly to the bacterial surface, as depicted in . Importantly, the statistical analysis revealed no significant difference in signal values (, p > 0.05). In summary, the synthesized SERS tags, especially those modified with 2.0 μg of WGA, demonstrated robust binding capabilities, as evidenced by the attachment of the tags to the bacterial surfaces.
Figure 3 Comparative optimization of WGA-SERS tags. (A) TEM images of S. aureus bound to WGA-SERS tags with various amounts of WGA. i~iv represent WGA concentrations of 0.25~2.5 μg. (B) The corresponding SERS intensities of SERS tags with S. aureus in various amounts of WGA. TEM images of sandwich composite structures formed by AuNSs or AuNPs-SERS tags with MGNPs adsorbed on S. aureus (C) and P. aeruginosa (D). Corresponding SERS signal measured from sandwich composite structures formed by AuNSs or AuNPs-SERS tags with S. aureus (E) and P. aeruginosa (F).

In this study, we conducted a comprehensive comparative analysis of SERS tags using conventional AuNPs and our proposed AuNSs as enhanced substrates for the detection of target bacteria, employing an equivalent concentration of WGA. TEM images presented in and demonstrate that within the MGNPs/bacteria/SERS tags complexes, a higher number of AuNSs adhered to S. aureus and P. aeruginosa compared to AuNPs. This observation is attributed to the heightened SERS-enhancing capability and surface extensibility of AuNSs. Specifically, the SERS tags utilizing AuNSs as the enhancement substrate exhibited a significant increase in Raman signals on the surfaces of MGNPs-enriched bacteria, as shown in and (p < 0.05). These findings emphasize the superior performance of SERS tags incorporating AuNSs@WGA in bacterial detection.
To assess the detection capability of the developed assay, bacterial samples of S. aureus and P. aeruginosa spanning a concentration range of 10–106 CFU/mL were meticulously prepared through gradient dilution. The implementation of the dual-identification SERS detection platform followed the proposed procedure, involving the separate incubation of MGNPs and bacteria, addition of equal amounts of WGA-SERS tags to form a sandwich structure, and the subsequent measurement of the Raman intensity at 1331 cm−1 of DTNB. As illustrated in and , the results for different concentrations of S. aureus and P. aeruginosa reveal a gradual increase in the intensity of the SERS signal with the rising concentration of bacteria in the sample. Calibration curves, depicted in and , were plotted by measuring the SERS signal intensity against the bacterial concentration of the two target bacteria. The SERS signal at Raman shift 1331 cm−1 exhibited a linear correlation with S. aureus and P. aeruginosa in the range of 10–106 CFU/mL (R2 = 0.987 and 0.985), respectively. Calculating the limit of detection (LOD) using the 3-fold standard deviation of the blank control determined the LOD for our SERS assay platform for S. aureus and P. aeruginosa to be 7 and 5 CFU/mL, respectively. The novel WGA-SERS tags, benefiting from the remarkable Raman enhancement effect of AuNSs and the efficient binding of WGA to pathogenic bacteria, offered stable and strong Raman signals. WGA, as a versatile bacterial recognition element, not only ensures strong bacterial binding affinity but also does so at a relatively low cost. In comparison to our previous work involving gold nanorods (AuNRs),Citation37 two-dimensional molybdenum disulfide (MoS2) nanosheets,Citation38 and graphene oxide (GO),Citation39 the proposed utilization of AuNSs as a novel SERS enhancement substrate introduces a simpler synthetic process and achieves superior SERS enhancement effects, enabling lower detection limits for the target bacteria. This strategic selection of SERS enhancement materials not only contributes to enhancing the stability and sensitivity of SERS tags but also broadens the potential application scope of bacterial detection.
Figure 4 Evaluation of the quantitative detection ability of the SERS biosensor. The recorded SERS spectra for sandwich complexes formed with S. aureus (A) and P. aeruginosa (B) in the concentration range of 10–106 CFU/mL. Corresponding calibration curves of the proposed SERS platform for S. aureus (C) and P. aeruginosa (D). Each spectrum is the average of three independently collected spectra.

Selectivity is a critical factor in determining the effectiveness of biosensors for practical applications. In this study, 105 CFU/mL of S. aureus or P. aeruginosa and other interfering strains (including L. mono, K. pneumoniae, A. baumannii and E. coli) were detected using our dual recognition SERS assay to evaluate the specificity of the developed platform. and show the corresponding SERS spectra at 1331 cm−1 that could be detected only in the presence of S. aureus or P. aeruginosa, while the SERS signal for the non-target bacteria was very weak. To investigate the utility and reliability of our proposed SERS assay platform in biological samples, the method’s recovery was evaluated by adding different concentrations of S. aureus and P. aeruginosa (5000, 500, 50 CFU/mL) to human urine samples. and show that very clear and stable SERS signals were generated for all urine samples spiked with S. aureus or P. aeruginosa. The recoveries ranged from 84.8% to 118.0%, with RSD below 11.42% (), showcasing the high reliability of the method in detecting biological samples. Therefore, our proposed biosensor demonstrates excellent applicability and accuracy for detecting S. aureus and P. aeruginosa.
Table 1 The Recovery Efficiency of S. aureus or P. aeruginosa in Human Urine Samples Using the Developed SERS Method
Conclusion
In conclusion, our study introduces a robust sandwich-structured SERS biosensor designed for the rapid and sensitive detection of S. aureus or P. aeruginosa. The incorporation of WGA-SERS tags, featuring WGA and DTNB-modified AuNSs, ensures strong anchoring to bacterial surfaces, facilitating stable Raman signal enhancement. Overcoming challenges associated with alternative recognition molecules, such as aptamers or antimicrobial peptides, our biosensor employs MNPs modified with specific antibodies as capture probes. The integration of magnetic separation of antibodies and the application of novel SERS tags significantly enhance the biosensor’s detection capability, achieving sensitivities as low as 7 CFU/mL for S. aureus and 5 CFU/mL for P. aeruginosa. With high speed, reproducibility, and specificity, our biosensor is well-suited for efficiently detecting bacteria in human urine samples. To the best of our knowledge, this work is the first to propose the combination of WGA and SERS tags for fabricating high-performance SERS tags in bacterial detection. By replacing the recognition element of the capture probe, the proposed universal SERS biosensor holds the potential for detecting various other pathogenic bacteria. Our biosensor stands out as a promising tool in pathogen detection and bioanalytical research, presenting a valuable contribution to the field. Moving forward, future research could explore further optimization and extension of this innovative SERS biosensor technology in different application scenarios. To achieve this, extending the detection range of this biosensor to target more pathogens for rapid and sensitive detection is a promising avenue. Additionally, considering further integration of intelligent technologies could enhance the operability and convenience of the biosensor in real-world scenarios, paving the way for more practical applications in various fields.
Abbreviations
S. aureus, Staphylococcus aureus; P. aeruginosa, Pseudomonas aeruginosa; WGA, Wheat germ agglutinin; Abs, Antibodies; AuNSs, Gold nanostars, DTNB, 5.5’-dithiobis(2-nitrobenzoic acid); magnetic gold nanoparticles (MGNPs); LOD, limit of detection; SERS, Surface-enhanced Raman scattering.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
We would like to express our sincere gratitude to Ping Yang, Nanjing Drum Tower Hospital, for his significant contributions to this research. Although not listed as a corresponding author, Ping Yang played a crucial role in data analysis and experimental design. His expertise and dedication greatly enriched the quality and depth of this study.
Additional information
Funding
References
- Atia AF, Afifi AF, Gaffer MMA, Elnahas NS, Oshiba SFA. Dendritic Cells as an Adjuvant to some Schistosoma mansoni Antigens for Vaccination in Experimental Schistosomiasis. Int J Curr Microbiol Appl Sci. 2020;9(3):32–53. doi:10.20546/ijcmas.2020.903.005
- Molehin AJ. Schistosomiasis vaccine development: update on human clinical trials. J Biomed Sci. 2020;27(1):28. doi:10.1186/s12929-020-0621-y
- Galvan DD, Yu Q. Surface-Enhanced Raman Scattering for Rapid Detection and Characterization of Antibiotic-Resistant Bacteria. Adv Healthc Mater. 2018;7(13):e1701335. doi:10.1002/adhm.201701335
- Someko H, Okazaki Y, Tsujimoto Y, Ishikane M, Kubo K, Kakehashi T. Diagnostic accuracy of rapid antigen tests in cerebrospinal fluid for pneumococcal meningitis: a systematic review and meta-analysis. Clin Microbiol Infect. 2023;29:310–319. doi:10.1016/j.cmi.2022.12.002
- Zhou Q, Natarajan B, Kannan P. Nanostructured biosensing platforms for the detection of food- and water-borne pathogenicEscherichia coli. Anal Bioanal Chem. 2023;415:3111–3129. doi:10.1007/s00216-023-04731-6
- Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID-19: a retrospective cohort study in a UK secondary-care setting. Clin Microbiol Infect. 2020;26(10):1395–1399. doi:10.1016/j.cmi.2020.06.025
- Lindner F, Diepold A. Optogenetics in bacteria - applications and opportunities. FEMS Microbiol Rev. 2022;46:undefined. doi:10.1093/femsre/fuab055
- Liu L, Zhang H, Xing S, et al. Copper-Zinc Bimetallic Single-Atom Catalysts with Localized Surface Plasmon Resonance-Enhanced Photothermal Effect and Catalytic Activity for Melanoma Treatment and Wound-Healing. Adv Sci. 2023;10(18):e2207342. doi:10.1002/advs.202207342
- Dina N, Tahir M, Bajwa S, Amin I, Valev V, Zhang L. SERS-based antibiotic susceptibility testing: towards point-of-care clinical diagnosis. Biosens Bioelectron. 2022;219:114843. doi:10.1016/j.bios.2022.114843
- Langer J, Jimenez de Aberasturi D, Aizpurua J, et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano. 2020;14:28–117.
- Huang X, Zhang Z, Chen L, et al. Multifunctional Au nano-bridged nanogap probes as ICP-MS/SERS dual-signal tags and signal amplifiers for bacteria discriminating, quantitative detecting and photothermal bactericidal activity. Biosens Bioelectron. 2022;212:114414. doi:10.1016/j.bios.2022.114414
- Lane LA, Qian X, Nie S. SERS Nanoparticles in Medicine: from Label-Free Detection to Spectroscopic Tagging. Chem Rev. 2015;115(19):10489–10529. doi:10.1021/acs.chemrev.5b00265
- Liu H, Gao X, Xu C, Liu D. SERS Tags for Biomedical Detection and Bioimaging. Theranostics. 2022;12(4):1870–1903. doi:10.7150/thno.66859
- Zhu A, Jiao T, Ali S, Xu Y, Ouyang Q, Chen Q. SERS Sensors Based on Aptamer-Gated Mesoporous Silica Nanoparticles for Quantitative Detection of Staphylococcus aureus with Signal Molecular Release. Anal Chem. 2021;93(28):9788–9796. doi:10.1021/acs.analchem.1c01280
- Zhuang J, Zhao Z, Lian K, et al. SERS-based CRISPR/Cas assay on microfluidic paper analytical devices for supersensitive detection of pathogenic bacteria in foods. Biosens Bioelectron. 2022;207:114167. doi:10.1016/j.bios.2022.114167
- Wu Z, Zhou J, Nkanga C, et al. One-Step Supramolecular Multifunctional Coating on Plant Virus Nanoparticles for Bioimaging and Therapeutic Applications. ACS Appl Mater Interfaces. 2022;14(11):13692–13702. doi:10.1021/acsami.1c22690
- Zhong Y, Zheng X, Li Q, Loh X, Su X, Zhao S. Antibody conjugated Au/Ir@Cu/Zn-MOF probe for bacterial lateral flow immunoassay and precise synergistic antibacterial treatment. Biosens Bioelectron. 2023;224:115033. doi:10.1016/j.bios.2022.115033
- Xu Y, He P, Ahmad W, et al. Catalytic hairpin activated gold-magnetic/gold-core-silver-shell rapid self-assembly for ultrasensitive Staphylococcus aureus sensing via PDMS-based SERS platform. Biosens Bioelectron. 2022;209:114240. doi:10.1016/j.bios.2022.114240
- Gao X, Yin Y, Wu H, et al. Integrated SERS Platform for Reliable Detection and Photothermal Elimination of Bacteria in Whole Blood Samples. Anal Chem. 2020;93(3):1569–1577. doi:10.1021/acs.analchem.0c03981
- Cao H, Gao Y, Jia H, et al. Macrophage-Membrane-Camouflaged Nonviral Gene Vectors for the Treatment of Multidrug-Resistant Bacterial Sepsis. Nano Lett. 2022;22(19):7882–7891. doi:10.1021/acs.nanolett.2c02560
- O’Neill E, Cosenza Z, Baar K, Block D. Considerations for the development of cost-effective cell culture media for cultivated meat production. Compr Rev Food Sci Food Saf. 2021;20(1):686–709. doi:10.1111/1541-4337.12678
- Shin W, Ahn G, Lee J-P, et al. Recent Advances in Engineering Aptamer-based Sensing and Recovery of Heavy Metals and Rare Earth Elements for Environmental Sustainability. Chem Eng J. 2023;472:144742. doi:10.1016/j.cej.2023.144742
- Mi F, Guan M, Hu C, Peng F, Sun S, Wang X. Application of lectin-based biosensor technology in the detection of foodborne pathogenic bacteria: a review. Analyst. 2021;146(2):429–443. doi:10.1039/D0AN01459A
- Skoll K, Palmetzhofer J, Lummerstorfer M, Anzengruber M, Gabor F, Wirth M. Human serum albumin nanoparticles as a versatile vehicle for targeted delivery of antibiotics to combat bacterial infections. Nanomedicine. 2023;50:102685. doi:10.1016/j.nano.2023.102685
- Masigol M, Fattahi N, Barua N, et al. Identification of Critical Surface Parameters Driving Lectin-Mediated Capture of Bacteria from Solution. Biomacromolecules. 2019;20(7):2852–2863. doi:10.1021/acs.biomac.9b00609
- Grossman AS, Escobar CA, Mans EJ, et al. A Surface Exposed, Two-Domain Lipoprotein Cargo of a Type XI Secretion System Promotes Colonization of Host Intestinal Epithelia Expressing Glycans. Front Microbiol. 2022;13:800366. doi:10.3389/fmicb.2022.800366
- Yang G, Meng X, Wang Y, Yan M, Aguilar ZP, Xua H. 2-Step lectin-magnetic separation (LMS) strategy combined with AuNPsbased colorimetric system for S. aureus detection in blood. Sens Actuators B. 2019;279:87–94. doi:10.1016/j.snb.2018.09.112
- Tu Z, Yang X, Dong H, et al. Ultrasensitive Fluorescence Lateral Flow Assay for Simultaneous Detection of Pseudomonas aeruginosa and Salmonella typhimurium via Wheat Germ Agglutinin-Functionalized Magnetic Quantum Dot Nanoprobe. Biosensors. 2022;12(11):942. doi:10.3390/bios12110942
- Zheng S, Yang X, Zhang B, et al. Sensitive detection of Escherichia coli O157:H7 and Salmonella typhimurium in food samples using two-channel fluorescence lateral flow assay with liquid Si@quantum dot. Food Chem. 2021;363:130400. doi:10.1016/j.foodchem.2021.130400
- Li J, Wang C, Shi L, et al. Rapid identification and antibiotic susceptibility test of pathogens in blood based on magnetic separation and surface-enhanced Raman scattering. Mikrochim Acta. 2019;186(7):475. doi:10.1007/s00604-019-3571-x
- Molina-Mora J, Fernando GG. Molecular Determinants of Antibiotic Resistance in the Costa Rican Pseudomonas aeruginosa AG1 by a Multi-omics Approach:A Review of 10 Years of Study. Phenomics. 2021;3:129–142. doi:10.1007/s43657-021-00016-z
- Zhou Z, Xiao R, Cheng S, et al. A universal SERS-label immunoassay for pathogen bacteria detection based on Fe3O4@Au-aptamer separation and antibody-protein A orientation recognition. Anal Chim Acta. 2021;1160:338421. doi:10.1016/j.aca.2021.338421
- Tu J, Wu T, Yu Q, et al. Introduction of multilayered magnetic core-dual shell SERS tags into lateral flow immunoassay: a highly stable and sensitive method for the simultaneous detection of multiple veterinary drugs in complex samples. J Hazard Mater. 2023;448:130912. doi:10.1016/j.jhazmat.2023.130912
- Liu X, Yang X, Li K, et al. Fe3O4@Au SERS tags-based lateral flow assay for simultaneous detection of serum amyloid A and C-reactive protein in unprocessed blood sample. Sens Actuators B. 2020;320:128350. doi:10.1016/j.snb.2020.128350
- Tu Z, Cheng S, Dong H, et al. Universal and ultrasensitive detection of foodborne bacteria on a lateral flow assay strip by using wheat germ agglutinin-modified magnetic SERS nanotags. RSC Adv. 2022;12(42):27344–27354. doi:10.1039/D2RA04735G
- Cheng S, Tu Z, Zheng S, et al. An efficient SERS platform for the ultrasensitive detection of Staphylococcus aureus and Listeria monocytogenes via wheat germ agglutinin-modified magnetic SERS substrate and streptavidin/ aptamer co-functionalized SERS tags. Anal Chim Acta. 2021;1187:339155. doi:10.1016/j.aca.2021.339155
- Wang J, Wu X, Wang C, et al. Facile synthesis of Au-coated magnetic nanoparticles and their application in bacteria detection via a SERS method. ACS Appl Mater Interfaces. 2016;8(31):19958–19967. doi:10.1021/acsami.6b07528
- Yu Q, Li J, Zheng S, et al. Molybdenum disulfide-loaded multilayer AuNPs with colorimetric-SERS dual-signal enhancement activities for flexible immunochromatographic diagnosis of monkeypox virus. J Hazard Mater. 2023;459:132136. doi:10.1016/j.jhazmat.2023.132136
- Wang C, Wang C, Li J, et al. Ultrasensitive and multiplex detection of four pathogenic bacteria on a bi-channel lateral flow immunoassay strip with three-dimensional membrane-like SERS nanostickers. Biosens Bioelectron. 2022;214:114525. doi:10.1016/j.bios.2022.114525