Abstract
Therapeutically relevant design of scaffolds is of special importance in the repair and regeneration of tissues including dentin and pulp. Here we exploit nanofiber matrices that incorporate bioactive glass nanoparticles (BGNs) and deliver the odontogenic drug dexamethasone (DEX) to stimulate the odontogenic differentiation of human dental pulp cells (HDPCs). DEX molecules were first loaded onto the BGN, and then the DEX-BGN complex was incorporated within the biopolymer nanofiber matrix through electrospinning. The release of DEX continued over a month, showing a slow releasing profile. HDPCs cultured on the DEX-releasing BGN matrices were viable, proliferating well up to 14 days. The odontogenic differentiation, as assessed by alkaline phosphatase activity, mRNA expression of genes, and mineralization, was significantly stimulated on the matrices incorporating BGN and further on those releasing DEX. The DEX-releasing BGN matrices highly upregulated the expression of the integrin subsets α1, α5, and β3 as well as integrin downstream signaling molecules, including focal adhesion kinase (FAK), Paxillin, and RhoA, and activated bone morphogenetic protein mRNA and phosphorylation of Smad1/5/8. Furthermore, the DEX-releasing BGN-matrices stimulated Akt and mammalian target of rapamycin (mTOR), which was proven by the inhibition study. Collectively, the designed therapeutic nanofiber matrices that incorporate BGN and deliver DEX were demonstrated to promote odontogenesis of HDPCs, and the integrins, bone morphogenetic protein, and mTOR signaling pathways are proposed to be the possible molecular mechanisms. While further in vivo studies are still needed, the DEX-releasing bioactive scaffolds are considered as a potential therapeutic nanomatrix for regenerative endodontics and tissue engineering.
Introduction
Biomatrices that have the therapeutic actions can lead to excellent outcome in the repair and regeneration of tissues, including dentin–pulp complexes.Citation1 The controlled and sustained delivery of therapeutic molecules to the target site of injuries and diseases possibly reduces inflammation, enhances angiogenesis, and accelerates odontogenic differentiation of dental pulp stem cells.Citation2–Citation5 Therefore, the design of matrices with therapeutically relevant actions is considered of special importance in improving the repair and regenerative potential of dentin and pulp tissues.
Over the last decades, many different forms of biomatrices have been developed in this area, including porous foams, macrochanneled scaffolds, and nanofibrous meshes. The latter, among others, have been attractive mainly due to their mimicking the structural feature of the native extracellular matrix at the nanoscale.Citation6–Citation9 Cells recognize the nanofibrous topology, adhere and spread well, and adopt their phenotypes properly depending on the surrounding biochemical cues.Citation5,Citation10 Our previous studies have shown that human dental pulp cells (HDPCs) actively grew on the nanofiber matrix made of biopolymers and differentiated actively into odontoblastic lineage.Citation11 Furthermore, when the nanofiber compositions were tailored to incorporate bioactive inorganic nanoparticles, such as those of calcium phosphates and bioglass, the odontogenic differentiation capacity could be substantially improved.Citation12
In this study, we aimed to utilize the nanofiber matrices and design them with therapeutic capacity through delivering drug molecules relevant for activating odontogenesis of dental stem cells. For this, we incorporated a bioactive nanoparticle composition, namely bioactive glass nanoparticles (BGNs), within biopolymer nanofiber matrices. In fact, BGN, as the bioactive inorganic nanocomponent, exerts excellent bone bioactivity, regulating the genetic profiles of osteoprogenitor/stem cells toward a lineage of bone-forming cells.Citation7,Citation8 Furthermore, as the therapeutic drug, we incorporated dexamethasone (DEX) within the nanofiber matrix.
In fact, DEX has been used as the bioactive molecule for bone tissue. Studies have shown that DEX treatment stimulated mesenchymal stem cell (MSC) proliferation and supported osteogenic lineage differentiation;Citation13 however, the direct use of DEX has been limited mainly due to its toxic side effects. For this reason, the use of DEX through incorporation into biomaterials is favored. When added to electrospun polycaprolactone (PCL) nanofibers, DEX was shown to increase the level of alkaline phosphatase (ALP) and mineralized matrix in human bone marrow MSCs.Citation14 Recently, nanofiber scaffolds were also designed to deliver DEX in concert with the bone morphogenetic protein 2 (BMP-2), which was shown to improve differentiation and bone repair.Citation15 Moreover, the proliferation and osteogenic differentiation of stem cells derived from periodontal ligament were stimulated by the long-term delivery of DEX that was incorporated within composite nanofibers.Citation16
Based on these series of studies, we proposed to utilize the DEX delivery system made of nano-biomatrices that have nanofibrous morphology and bioactive nanocomponent within the composition, targeting odontogenic differentiation of dental stem cells. We cultured dental stem cells derived from human pulp (HDPCs) upon the DEX-delivering nano-biomatrices and investigated the odontogenesis of cells; moreover, we addressed the biological signaling pathways behind these events.
Materials and methods
Nano-biomatrices and DEX loading
All experiments were approved by the ethics committee of Kyung Hee University (Seoul, Republic of Korea). First, the nanocomponent (BGN) added within the nanofiber matrix was prepared by the sol–gel approach.Citation16,Citation17 In brief, 5 g polyethylene glycol (PEG) was dissolved in 120 mL methanol with pH adjusted to 12.5, and then 0.299 g Ca (NO3)2·4H2O was added and dissolved. Separately, 0.793 g TEOS (tetraethyl orthosilicate) was added to 30 mL methanol, which was then added dropwise to the pH-adjusted solution with ultrasonication. A white precipitate was obtained with 24 hours of stirring, which was then separated and washed with a water/ethanol mixture thoroughly and then dried overnight. To remove the PEG template, the powder was calcined at 600°C for 5 hours to obtain BGNs. The BGN powder was then amine-functionalized with 3-aminopropyl triethoxysilane at 80°C for 12 hours.
For the loading of DEX (C22H28FNa2O8P, a phosphate form in disodium salt), the BGN was dispersed at 10 mg/mL in distilled water containing DEX at different concentrations (5 mg/mL, 10 mg/mL, 15 mg/mL, 20 mg/mL, 25 mg/mL, and 30 mg/mL). The amounts of DEX loaded were determined by the absorbance at 242 nm using Ultraviolet–visible (UV–vis) spectrometry (Libra S22, Biochrom). The release of DEX from the BGN was also recorded by adding 10 mg of DEX-loaded BGN in 50 mL distilled water at 37°C for different time periods. At predetermined time points, 1 mL of test sample was withdrawn and the release of DEX was monitored by the UV–vis measurement.
For the generation of nanofiber matrices made of DEX-loaded BGN and biopolymers, the electrospinning technique was used. The polymer solution was composed of 10 wt% gelatin/10 wt% PCL dissolved in TFE (trifluoroethanol) at 40°C, to which the BGN (or BGN-DEX) was added at 2.5 wt% with respect to the polymer. The composite solutions were then electrospun at an injection rate of 1 mL/h, voltage 10.5 V, and distance 14.5 cm to produce nanofibrous nonwoven meshes. The prepared samples were examined for the nanostructure morphologies by scanning electron microscopy (SEM, JSM-6510; JEOL, Tokyo, Japan) and transmission electron microscopy (TEM, JEM-3010; JEOL).
The degradation of the composite nanofiber was examined by immersing the nanofiber sample in 10 mL phosphate-buffered saline (pH =7.4) at 37°C. After immersion for different time periods (7 days, 14 days, 21 days, and 28 days), the sample was taken out, dried, and the weight change was recorded. The release of DEX from the biomatrices was investigated by high-performance liquid chromatography (ACME 9000 system; Younglin Instrument, Ltd., Republic of Korea). Fifty milligrams of the nanofiber mesh was immersed in 10 mL distilled water and incubated at 37°C. At predetermined time points, 1 mL of the release medium was sampled for the DEX analysis and the medium was refreshed.
Cell culture and proliferation
Immortalized HDPCs, transfected with human telomerase catalytic component, were kindly provided by Professor Takashi Takata (Hiroshima University, Japan).Citation18 Cells were cultured in α-minimum essential medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin in a humidified atmosphere of 5% CO2 at 37°C. In order to induce odontogenesis and to study the effects of DEX release from the nanofiber matrices, the cell culture medium was composed of either without DEX (negative control) or with DEX (positive control), ie, α-minimum essential medium supplemented with 10% fetal bovine serum, 50 mg/mL ascorbic acid, and 10 mM glycerophosphate, with or without 0.1 µM DEX. The culture medium was replaced every 2 days during the incubation period.
During culture for up to 14 days, the cell viability was assayed by means of a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTS) assay kit (Cell Titer 96 Aq Non-Radioactive Cell Assay; Promega Corporation, Fitchburg, WI, USA). Briefly, MTS was added to the wells and incubated for 3 hours. The blank used was the culture medium with MTS. The absorbance was measured at 492 nm in an enzyme-linked immunosorbent assay 96-well plate reader. Cell migration was monitored using quartz crystal microbalance (QCM) chemotaxis in 24-well colorimetric cell migration kits (Chemicon, Temecula, CA, USA) following the manufacturer’s instructions. Measurements were done in triplicate, and the results were expressed as the mean in each experiment.
Odontogenic differentiation assays
The odontogenic differentiation of HDPCs induced by the DEX-releasing nano-biomatrices was assessed by the mRNA expressions, ALP activity, and mineralization. For the reverse transcription polymerase chain reaction (RT-PCR) analysis, first the RNA of the cells was extracted using the Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Then 1 mg of RNA isolated from each culture was reverse-transcribed using oligo (dT) primers (Hoffman-La Roche Ltd., Basel, Switzerland) with an AccuPower RT PreMix (Bioneer Corporation, Daejeon, South Korea). Thereafter, 60 ng of cDNA was amplified with 1 mM primer. The primer sequences and PCR conditions are given in . PCR products were subjected to electrophoresis on 1.5% agarose gels stained with ethidium bromide.
Table 1 Reverse transcription polymerase chain reaction (RT-PCR) primers and conditions
The ALP activity was performed in assay buffer (10 mM MgCl2 and 0.1 M alkaline buffer, pH 10.3) containing 10 mM p-nitrophenylphosphate as a substrate. Absorbance was measured at 410 nm using an enzyme-linked immunosorbent assay reader.
For the mineralization assay, the Alizarin Red S (ARS) staining method was used. For this, the treated cells were washed thoroughly with distilled water and then fixed in 10% formalin for 30 minutes at 4°C. Cells were incubated in 40 mM ARS regent (Sigma-Aldrich Co., St Louis, MO, USA) for 20 minutes at room temperature followed by complete washes with distilled water, and then observed under a light microscope (Olympus Corporation, Tokyo, Japan).
Mechanistic study on signaling pathways
The signaling mechanisms behind the odontogenic differentiation were investigated in terms of integrins/downstream signals, MAPK pathways, BMP signaling, and Akt pathways. For integrin signaling, the mRNA levels of some integrin subsets representative for HDPCs (α5, α1, β1, and β3) were first analyzed. Furthermore, the integrin downstream signals were identified by the expressions of the phosphorylated form of intracellular adhesive molecules, including focal adhesion kinase (FAK), Paxillin, and RhoA, by means of Western blotting analysis. For BMP signaling, the mRNA levels of BMP-2 and BMP receptors (BMPRa-1, BMPRa-2, and BMPR2) were examined, and the protein expression of p-Smad1/5/8 was also analyzed. In particular, the p-Smad1/5/8 expression was examined by means of the Noggin blocking test. For the Akt/mammalian target of rapamycin (mTOR) signaling study, the expressions of p-Akt, p-70S6K, and p-4EBP1 were analyzed by Western blot, and the blocking study using rapamycin (RAPA) was also carried out.
The conditions for RT-PCR analysis were similar to those used for the odontogenic study, and the primer sequences and PCR conditions were as detailed in . For Western blotting, the cell extracts were prepared by solubilizing cells in the protein lysis buffer (5 mM ethylenediaminetetraacetic acid [EDTA], 1 mM MgCl2, 50 mM Tris–HCl [pH 7.5], 0.5% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, and 1 mM N-ethylmaleimide) for 30 minutes on ice. Fifty micrograms of the protein was separated by sodium dodecyl sulfate polyacrylamide electrophoresis and electro-transferred to polyvinylidene difluoride membranes. The membranes were treated with blocking solution for 1 hour and were incubated with 1:1,000 dilution primary antibody against phospho-focal adhesion kinase (p-FAK; Santa Cruz Biotechnology Inc., Dallas, TX, USA), p-Paxillin (Santa Cruz Biotechnology Inc.), actin (Santa Cruz Biotechnology Inc.), Rho-A (Cell Signaling Technology, Danvers, MA, USA), p-Smad1/5/8 (Cell Signaling Technology), p-p38 (Cell Signaling Technology), and p-Akt (Cell Signaling Technology). The blots were followed by incubation for 1 hour with the appropriate horseradish peroxidase-conjugated secondary antibody. The proteins were then visualized using an enhanced chemiluminescence system (Amersham, Piscataway, NJ, USA).
Statistical analysis
Data are presented as mean and SD. Statistical analysis was performed by one-way analysis of variance followed by multiple-comparison Tukey’s test using SPSS Version 22.0 (IBM Corporation, Armonk, NY, USA). Statistical significance was considered at P<0.05.
Results and discussion
Nano-biomatrices incorporate bioactive nanoparticles and release DEX drug sustainably
The nano-morphology of the BGN used for the delivery of DEX and also for a nano-additive to polymer matrices was examined by TEM. Spherical nanoparticles with a uniform size of 63 nm ± 12.3 were developed well, and a closer examination revealed the mesopores to be evenly distributed in the nanoparticles (). After the loading with DEX, the TEM image of samples was also revealed (). The mesoporous image appeared diffused due to the pore-filling of DEX molecules. The loading amount of DEX in BGN samples was then monitored when 10 mg/mL of BGN was used while DEX concentration varied up to 40 mg/mL (). DEX loading amount increased gradually with increasing DEX concentration used initially, and then saturated to 630 µg of DEX which gave as high as 63% of loading efficiency. The release of DEX from the BGN samples was then recorded (). The DEX release was highly abrupt, showing ~80% release within 24 hours followed by an almost complete release within 3 days.
Figure 1 Characteristics of DEX-loaded BGN and the composite nanofibers.
Notes: (A and B) TEM of BGN at different magnifications. (C) DEX adsorption amount onto BGN, recorded at varying BGN loading concentrations. (D) DEX release amount from the BGN, showing a rapid release pattern. (E) SEM of composite nanofiber. (F) TEM internal image of the nanofiber. (G) Degradation of the nanofiber measured via weight change up to 28 days. (H) DEX release from the composite nanofiber, showing a highly sustained release pattern.
Abbreviations: DEX, dexamethasone; BGN, bioactive glass nanoparticle; TEM, transmission electron microscopy; SEM, scanning electron microscopy; h, hours; wt, weight.
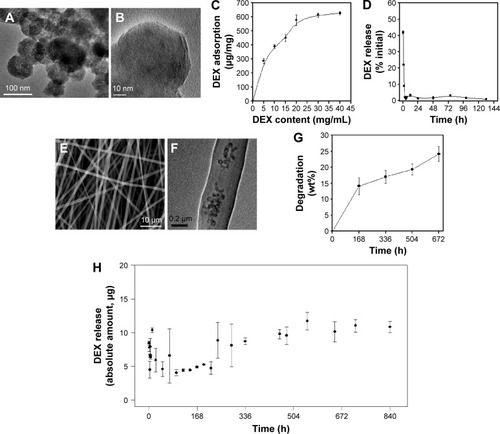
Next, the DEX-loaded BGNs were incorporated within the PCL–gelatin nanofiber matrix to produce a nanofibrous mesh through electrospinning. 2.5 wt% of the DEX-BGN complex was loaded within the nanofiber, and the DEX loading amount used was below the saturation point (0.286 mg DEX per 1 mg BGN). The SEM images of the composite nanofibers showed the production of nonwoven mesh of fibrous morphology (). On the TEM image, the internal morphology of the nanofiber was revealed, where the nanospherical BGNs were dispersed well within the biopolymer nanofiber matrix (). The degradation of the composite nanofiber was then examined. The weight loss of the nanofiber during immersion in a saline solution was ~15% for the first 7 days and then ~23% for up to 28 days ().
We further investigated the release of DEX from the produced DEX-BGN-incorporated nanofiber matrices. The DEX amount released at each time point was recorded for up to 35 days (). Interestingly, the released DEX amount was almost similar up to the period tested, except the initial rapid release (10 hours). This initial burst release was mainly due to the DEX molecules present in the biopolymer matrix that might have been pre-released during the preparation of the composite solution and nanofibers. More importantly, the continued release of DEX over a couple of weeks (and even more possibly) at almost constant dose is considered of great merit in long-term drug delivery systems.Citation19–Citation21 From a practical aspect, this DEX-releasing system would be highly beneficial for providing a constant-dosed odontogenic drug to cells during the long-term culture period, without the supplement of the drug within a culture medium; further, this sustained and controlled DEX-releasing system will be potentially useful for in vivo implantable scaffolds where the DEX drug released at a constant dose can favorably act in the surrounding cells for tissue (dentin–pulp) regeneration. This release profile of DEX might be the result of combined actions of the diffusion of DEX and the degradation of biopolymer matrices. If the DEX molecules were released from nondegradable matrix, the release pattern might follow a certain power-law pattern; however, the hydrolytic degradation of the biopolymer matrix might accelerate the DEX release, ultimately leading to an almost linear release pattern. After confirming the release of DEX, we next sought to investigate the effects of DEX-releasing nanofiber matrices on the behavior of HDPCs.
DEX-delivering bioactive nano-biomatrices stimulate odontogenesis of HDPCs
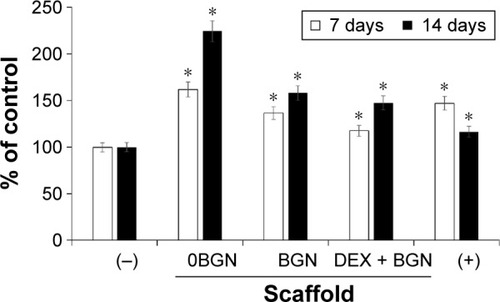
Before addressing the odontogenic differentiation of cells, we examined the viability of cells upon the DEX-releasing nanofiber matrices. The culture medium without DEX(−) or with DEX(+) was used as negative and positive control, respectively; moreover, nanofiber matrices without BGN (0BGN), or with BGN only (BGN), or DEX-loaded BGN (DEX-BGN) were used as experimental groups involving scaffolds. The cell viability was measured at day 7 and day 14 of culture, using an MTT assay, as presented in . Cells were highly proliferative on day 7 on the nanofiber matrices, with the highest level on 0BGN and the lowest level on DEX-BGN; however, both were still higher than that in DEX(−) control. At day 14, the cell viability increased substantially on the 0BGN matrix; but on other matrices the cell proliferation was not as much, with the DEX(+) control showing the lowest level. While the cell viability on the nano-biomatrices were up- and downregulated with time, this was closely related to the time-dependent cell differentiation as well, making the phenomenon quite complicated.
Figure 2 Cell viability measured by MTT assay.
Notes: DEX-free (−) and DEX-containing (+) media were used as negative and positive control, respectively. Data are the representatives of three independent experiments. *P<0.05 vs (−) control.
Abbreviations: DEX, dexamethasone; BGN, bioactive glass nanoparticle.
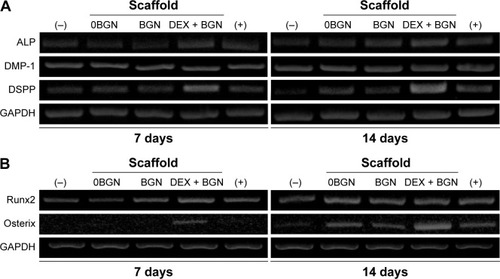
We next investigated the odontogenic differentiation behaviors of HDPCs. First, the mRNA expressions of odontoblastic marker genes (ALP, DSPP, and DMP-1) were assessed by RT-PCR (). In particular, the DSPP and DMP-1 genes are known to be identifying markers of odontoblasts, and play crucial roles in the early odontoblast differentiation and late dentin mineralization of dental pulp cells.Citation22,Citation23 From the RT-PCR bands, the DEX-BGN group, compared to others, showed higher expressions of genes, and particularly at day 14 for DSPP gene. Further, we analyzed the transcription factors Runx2 and Osterix, which are the major genes highly expressed during osteogenic/osteogenic induction in various cell types.Citation24 There were substantially higher expressions of the genes (particularly Osterix) when cultured on the DEX-BGN matrix ().
Figure 3 mRNA expression of genes.
Notes: (A) Odontogenic marker genes (ALP, DMP-1, DSPP). (B) Transcriptional factors involved in odontogenesis (Runx2 and Osterix).
Abbreviations: ALP, alkaline phosphatase; DMP-1, dentin matrix protein-1; DSPP, dentin sialophosphoprotein; DEX, dexamethasone; BGN, bioactive glass nanoparticle; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
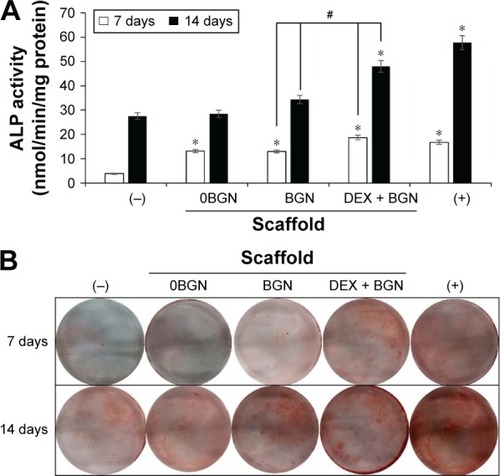
We further analyzed the odontogenic differentiation of HDPCs by the ALP (). The ALP level increased with the culture time, implying the time-dependent increase of ALP during odontogenic differentiation, a characteristic phenomenon widely reported elsewhere.Citation25 Compared to DEX(−) control, all the matrix groups showed significantly higher ALP levels. Moreover, the DEX-BGN group had significantly higher ALP level than BGN only, demonstrating the role of DEX released from the matrix. ARS staining was then performed to identify cellular mineralization. Cells cultured for 7 days and 14 days were treated with ARS dye, and the reaction products were gathered and visualized under optical microscopy (). Red stained images became much clearer with increasing time, and, furthermore, the matrix groups showed more stained images than DEX(−) control, while DEX(+) control exhibited the most. Compared to BGN only, the DEX-BGN group appeared to stain more.
Figure 4 ALP activity and ARS staining.
Notes: (A) ALP activity, and (B) ARS staining image, showing the odontogenic differentiation of HDPCs. Data are the representatives of three independent experiments. *P<0.05 vs (−) control; #P<0.05 in comparison between BGN and DEX-BGN.
Abbreviations: ALP, alkaline phosphatase; ARS, Alizarin Red S; HDPC, human dental pulp cell; BGN, bioactive glass nanoparticle; DEX, dexamethasone.
Collectively, the series of assays on odontogenic differentiation of HDPCs clearly demonstrated the effective role of DEX incorporated within the nanofiber matrix. In particular, the DEX-BGN group showed a level almost comparable to that of DEX(+) control, ie, the cells cultured on the in situ DEX-releasing matrix, which contained DEX-free medium, behaved similar to those cultured with DEX-maintaining medium in dish. Although this in vitro culture condition can signify the similar effectiveness of the two systems, the DEX-releasing matrix system has the unique merit for the in vivo implantations where additional DEX cannot be supplemented externally. Interestingly, all matrix groups (even BGN- and DEX-free) stimulated the odontogenesis of HDPCs when the results were compared with DEX(−) control; in other words, the nanofiber matrix culture itself had some effective role in odontogenic differentiation under the DEX-free culture medium. In fact, the nanotopological cues have often been shown to activate osteoblast activity of MSCs.Citation26 For the case of HDPCs, nanotopological substrates including biopolymer nanofibers and calcium phosphate nano-grained cements have recently been demonstrated to have similar effects on odontogenic differentiation.Citation27–Citation29 Furthermore, our odontogenesis results showed that the BGN-incorporated nanomatrix was even better than BGN-free nanomatrix, signifying the bioactive nanocomponent role of BGN. Our previous studies have shown that the BGN added to biopolymeric matrices stimulates odontogenesis of HDPCs.Citation11,Citation12 While the results of this study clearly demonstrate the beneficial action of DEX molecules released from the nanofiber matrices in the stimulation of odontogenesis, possible side effects of the DEX drug might also be of concern because DEX, as a powerful glucocorticoid, has been shown to exert adverse effects when used at high doses and for long-term therapy. However, in this study, we designed a system that allows local delivery of small doses of DEX over months, which is considered to relieve the concern that has been raised primarily in the systemic delivery systems. In fact, many local delivery systems of DEX have been reported to stimulate cellular responses in vitro and/or to improve tissue healing in vivo with little side effects.Citation30–Citation33 Even so, future in vivo studies may be needed to confirm this aspect of the current delivery system.
Integrin, BMP, and Akt/mTOR signaling pathways are involved in the events
After confirming the stimulating role of the DEX-releasing bioactive nanofiber matrices in odontogenic differentiation, we next sought to address the signal transduction mechanisms involved in the events. First, we focused on integrin and its downstream pathway, because this has been considered as one of the most important pathways in the matrix-related cell differentiation behaviors.Citation34,Citation35 The mRNA expressions of the integrin subsets α1, α5, β1, and β3 were analyzed via RT-PCR (). The mRNA levels of integrins α1, α5, and β3 were upregulated mainly by the BGN and DEX-BGN groups, whereas integrin β1 appeared not to be affected. Integrins are the major adhesion receptors that mediate cell adhesion and attachment,Citation34 and different combinations of integrin subsets are expressed depending on the underlying substrates and ligands. The integrin subsets activated in our study (α2, α5, β1) are known to be associated with collagen binding, which are thus considered to be present at greater levels over the matrices used herein, particularly BGN-containing ones. Our previous studies on bioactive scaffolds incorporating BGN have also shown similar results on the integrin expressions.Citation11,Citation12
Figure 5 Integrin signaling pathway analyses.
Notes: (A) mRNA expressions of integrin subsets (α1, α5, β1, and β3), by RT-PCR. GAPDH was used as the housekeeping gene. (B) Protein expressions of intracellular adhesive molecules (p-Paxillin, p-FAK, and RhoA), by Western blot. β-actin was used as the control.
Abbreviations: RT-PCR, reverse transcription polymerase chain reaction; p-FAK, phospho-focal adhesion kinase; BGN, bioactive glass nanoparticle; DEX, dexamethasone; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
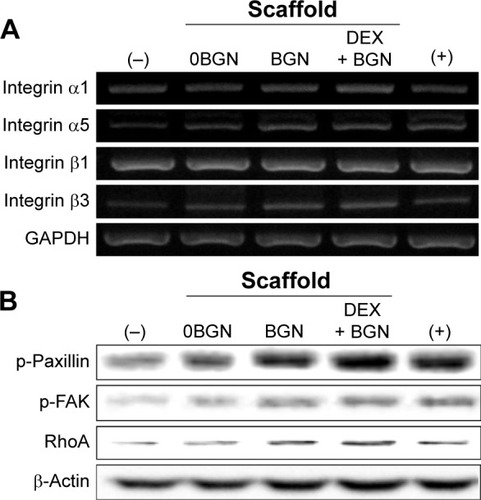
Next, the integrin downstream signaling molecules, including FAK, Paxillin, and RhoA, were analyzed by Western blotting (). The expressions of phosphorylated-Paxillin and p-FAK and total RohA were highly activated with the culture on the nanofiber matrices, and more substantial activation of proteins was revealed on BGN and DEX-BGN. Engagement of cell surface integrins is associated with rapid tyrosine phosphorylation of several intracellular adhesive molecules, such as FAK and Paxillin, which are two focal adhesion-associated proteins that function in transmitting signals downstream of integrins.Citation35 Moreover, RhoA, the most extensively studied Rho family member, is known to be activated by the integrins.Citation36 Therefore, the significant stimulation of those intracellular adhesive molecules by the BGN and DEX-BGN suggests that integrin and its outside-in signaling pathways are heavily involved in the matrices-based HDPC differentiation behaviors.
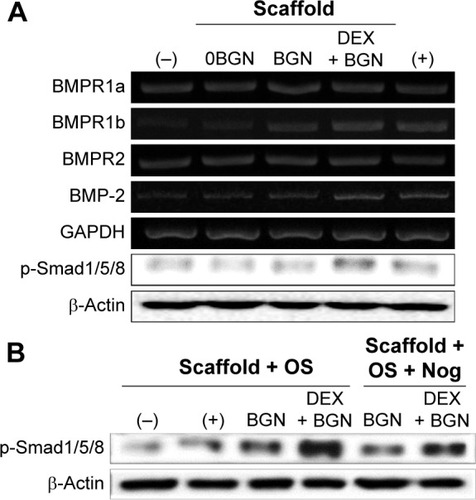
We next focused on the BMP signaling pathway. The mRNA expressions of BMP-2 and key BMP receptors (BMPR1a, BMPR1b, and BMPR2) were upregulated by the DEX-BGN group. Furthermore, analysis of the phosphorylation of Smad1/5/8 revealed substantial activation on the DEX-BGN (). To verity the matrix effects on the BMP pathway, the BMP inhibition study was further carried out. For this, cells were pretreated with a culture medium with or without the BMP inhibitor Noggin. The highly activated phosphorylation of Smad1/5/8 by DEX-BGN was significantly inhibited by the treatment with Noggin (). The results clearly demonstrated that the BMP signaling pathway is critically involved in the behavior of HDPCs upon the DEX-delivering bioactive nanofiber matrices.
Figure 6 BMP-2 signaling pathway analyses.
Notes: (A) mRNA levels of BMP receptors (R1a, R1b, and R2) and BMP-2, as well as the protein expression of p-Smad1/5/8. (B) Inhibition study to verify the signaling pathway, where a BMP inhibitor, Noggin (10 µg/mL) was used to treat the cells for 1 hour, and then cultured for 2 days before Western blot analysis. Data are representative of three independent experiments.
Abbreviations: BMP, bone morphogenetic protein; BGN, bioactive glass nanoparticle; DEX, dexamethasone; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Nog, noggin.
In fact, the BMP signaling pathway is known to play a vital role in osteogenic differentiation as well as in bone development and regeneration.Citation37 More recently, it was reported that integrin overlapping with BMP-2 receptors plays an essential role in the BMP-2 induction of osteoblast differentiation.Citation38 Moreover, the activation of surface integrin of MSCs, which is influenced by the extracellular matrix’s elasticity, in turn, can modulate BMP receptor internalization, thereby regulating the lineage specification of MSCs.Citation39 Our results also demonstrated that the DEX-releasing nano-biomatrix cues activated the expressions of integrins in HDPCs, which could in turn possibly stimulate the BMP signal transduction mechanism.
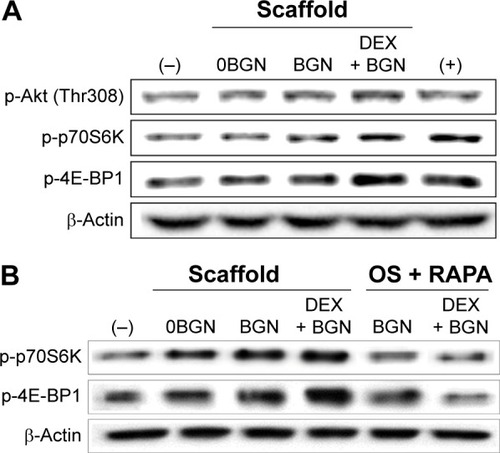
We next focused on the mTOR pathway, as this is known to be one of the principal signaling cascades regulating osteoblastic/odontoblastic differentiation.Citation40 The phosphorylated form of Akt and mTOR targeting p70S6 kinase (p70S6K) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) were shown to be highly upregulated in the matrix groups, particularly in the DEX-BGN matrix (). mTOR, a major downstream target of PI3K/Akt, is known to regulate various cellular functions required for cell growth and proliferation by phosphorylating a series of substrates, including p70S6K and 4E-BP1.Citation40 To clarify the effects in the Akt/mTOR signaling, we tested again the mTOR-inhibition experiment. The cells were pretreated with RAPA, a selective mTOR inhibitor. RAPA, a well-known inhibitor of mTOR, first forms a complex with a 12 kDa cytosolic protein designated FK-506-binding protein 12, and this complex binds mTOR and inhibits its function.Citation41 After the RAPA treatment, there was significant attenuation of p-p70S6K and p-4E-BP1, which was more noticeable in the DEX-BGN matrix ().
Figure 7 Akt/mTOR signaling pathway, analyzed by Western blot.
Notes: (A) Analysis of p-Akt, p-p70S6K, and p-4E-BP1. (B) Inhibition study to verify the signaling pathway, where cells were pretreated with rapamycin (100 ng/mL) for 1 hour and then incubated with samples for 30 minutes. Data are representative of three independent experiments.
Abbreviations: BGN, bioactive glass nanoparticle; DEX, dexamethasone; mTOR, mammalian target of rapamycin; RAPA, rapamycin.
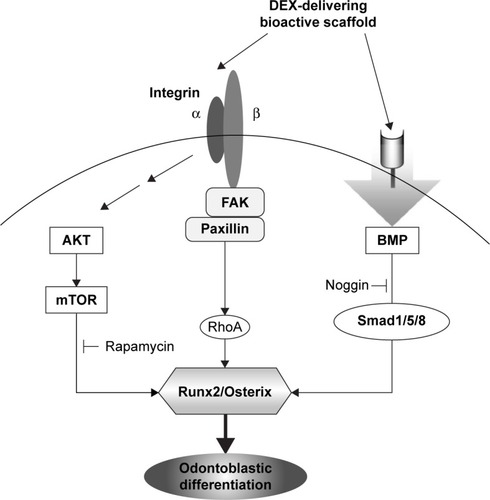
It is known that BMP signaling is linked to the mTOR pathway. BMP expression has been shown to activate P70S6K, a downstream effector of mTOR, suggesting that BMP-induced osteogenesis is mediated by mTOR activation.Citation42 Furthermore, integrin signaling is also closely related with the Akt/mTOR intracellular signaling.Citation43 Suppression of PI3K and mTOR signaling using inhibitors such as LY294002, wortmannin, and RAPA has been shown to promote osteogenesis in vitro.Citation44–Citation46 Therefore, the integrin and BMP stimulations, which are influenced by the DEX-releasing bioactive substrates, might trigger the Akt/mTOR transduction signaling pathway. is a schematic diagram showing the signaling pathways of integrins, BMP, and Akt/mTOR, which are possibly involved in the odontogenic stimulation of HDPCs, upon the culture with DEX-releasing bioactive nanofiber matrices.
Figure 8 Schematic illustration showing the integrin, BMP, and mTOR signaling pathways triggered by the exposure to DEX-delivering bioactive nanofiber matrices, which ultimately stimulated the odontogenic differentiation of HDPCs.
Abbreviations: BMP, bone morphogenetic protein; DEX, dexamethasone; mTOR, mammalian target of rapamycin; HDPC, human dental pulp cell; FAK, focal adhesion kinase.
Conclusion
Bioactive nanofiber matrices developed to release DEX and incorporate BGN were demonstrated to stimulate odontogenic differentiation of HDPCs. The activation of integrin, BMP/Smad, and Akt/mTOR signaling pathways are the possible key mechanisms involved in the events. While more in-depth studies on in vivo applicability are necessary, the developed nanomatrices can be a potential therapeutic platform for the regeneration of dentin–pulp complex tissues.
Acknowledgments
This study was supported by the grants Priority Research Centers Program No 2009-0093829, Global Research Lab Program No 2015032163, and MRC Program No 2012R1A5A2051384 of the National Research Foundation, Republic of Korea.
Disclosure
The authors report no conflicts of interest in this work.
References
- SharmaSSikriVSharmaNKSharmaVMRegeneration of tooth pulp and dentin: trends and advancesAnn Neurosci20101713143
- AboutIBotteroMJde DenatoPCampsJFranquinJCMitsiadisTAHuman dentin production in vitroExp Cell Res20002581334110912785
- BohlKSShonJRutherfordBMooneyDJRole of synthetic extracellular matrix in development of engineered dental pulpJ Biomater Sci Polym Ed1998977497649686338
- HoltgraveEADonathKResponse of odontoblast-like cells to hydroxyapatite ceramic granulesBiomaterials19951621551597734651
- OhgushiHCaplanAIStem cell technology and bioceramics: from cell to gene engineeringJ Biomed Mater Res199948691392710556859
- KimHWKimHEKnowlesJCProduction and potential of bioactive glass nanofibers as a next-generation biomaterialAdv Funct Mater2006161215291535
- KimHWLeeHHChunGSBioactivity and osteoblast responses of novel biomedical nanocomposites of bioactive glass nanofiber filled poly(lactic acid)J Biomed Mater Res A200885365166317876800
- KimHWSongJHKimHEBioactive glass nanofiber-collagen nanocomposite as a novel bone regeneration matrixJ Biomed Mater Res A200679369870516850456
- SongJHKimHEKimHWCollagen-apatite nanocomposite membranes for guided bone regenerationJ Biomed Mater Res B Appl Biomater200783124825717385223
- PrescottRSAlsaneaRFayadMIIn vivo generation of dental pulp-like tissue by using dental pulp stem cells, a collagen scaffold, and dentin matrix protein 1 after subcutaneous transplantation in miceJ Endod200834442142618358888
- BaeWJMinKSKimJJKimJJKimHWKimECOdontogenic responses of human dental pulp cells to collagen/nanobioactive glass nanocompositesDent Mater201228121271127923031484
- KimGHParkYDLeeSYOdontogenic stimulation of human dental pulp cells with bioactive nanocomposite fiberJ Biomater Appl201529685486625098335
- JaiswalNHaynesworthSECaplanAIBruderSPOsteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitroJ Cell Biochem19976422953129027589
- MartinsADuarteARFariaSMarquesAPReisRLNevesNMOsteogenic induction of hBMSCs by electrospun scaffolds with dexamethasone release functionalityBiomaterials201031225875588520452016
- LiLZhouGWangYYangGDingSZhouSControlled dual delivery of BMP-2 and dexamethasone by nanoparticle-embedded electrospun nanofibers for the efficient repair of critical-sized rat calvarial defectBiomaterials20153721822925453952
- El-FiqiAKimJHKimHWOsteoinductive fibrous scaffolds of biopolymer/mesoporous bioactive glass nanocarriers with excellent bioactivity and long-term delivery of osteogenic drugACS Appl Mater Interfaces2015721140115225531645
- El-FiqiAKimHWMesoporous bioactive nanocarriers in electrospun biopolymer fibrous scaffolds designed for sequential drug deliveryRSC Adv20144944444452
- KitagawaMUedaHIizukaSImmortalization and characterization of human dental pulp cells with odontoblastic differentiationArch Oral Biol200752872773117350587
- JayantRDMcShaneMJSrivastavaRPolyelectrolyte-coated alginate microspheres as drug delivery carriers for dexamethasone releaseDrug Deliv200916633134019606947
- PatilSDPapadmitrakopoulosFBurgessDJConcurrent delivery of dexamethasone and VEGF for localized inflammation control and angiogenesisJ Control Release20071171687917169457
- WangQWangJXLuQHDetamoreMSBerklandCInjectable PLGA based colloidal gels for zero-order dexamethasone release in cranial defectsBiomaterials201031184980498620303585
- HanksCTFangDSunZEdwardsCAButlerWTDentin-specific proteins in MDPC-23 cell lineEur J Oral Sci1998106suppl 12602669541235
- NarayananKSrinivasRRamachandranAHaoJQuinnBGeorgeADifferentiation of embryonic mesenchymal cells to odontoblast-like cells by overexpression of dentin matrix protein 1Proc Natl Acad Sci U S A20019884516452111287660
- NakashimaKZhouXKunkelGThe novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formationCell20021081172911792318
- HarrisHThe human alkaline phosphatases: what we know and what we don’t knowClin Chim Acta199018621331502178806
- ShiXWangYVarshneyRRRenLGongYWangDAMicrosphere-based drug releasing scaffolds for inducing osteogenesis of human mesenchymal stem cells in vitroEur J Pharm Sci2010391–3596719895885
- KimJJBaeWJKimJMMineralized polycaprolactone nanofibrous matrix for odontogenesis of human dental pulp cellsJ Biomater Appl20142871069107823839784
- LeeSKLeeSKLeeSIEffect of calcium phosphate cements on growth and odontoblastic differentiation in human dental pulp cellsJ Endod20103691537154220728723
- ZhangJParkYDBaeWJEffects of bioactive cements incorporating zinc-bioglass nanoparticles on odontogenic and angiogenic potential of human dental pulp cellsJ Biomater Appl201529795496425201908
- DingSLiJRLuoCLiLYangGZhouSBSynergistic effect of released dexamethasone and surface nanoroughness on mesenchymal stem cell differentiationBiomater Sci201311010911100
- ShresthaSDiogenesAKishenATemporal-controlled dexamethasone releasing chitosan nanoparticle system enhances odontogenic differentiation of stem cells from apical papillaJ Endod20154181253125825956605
- YangCMJiangLQBuSJIntravitreal administration of dexamethasone-loaded PLGA-TPGS nanoparticles for the treatment of posterior segment diseasesJ Biomed Nanotechnol2013991617162323980509
- ZhangLHLiYZhangCWangYSSongCXPharmacokinetics and tolerance study of intravitreal injection of dexamethasone-loaded nanoparticles in rabbitsInt J Nanomedicine2009417518319774116
- HynesROIntegrins: bidirectional, allosteric signaling machinesCell2002110667368712297042
- SchallerMDFAK and paxillin: regulators of N-cadherin adhesion and inhibitors of cell migration?J Cell Biol2004166215715915263014
- MoghePVBerthiaumeFEzzellRMTonerMTompkinsRGYarmushMLCulture matrix configuration and composition in the maintenance of hepatocyte polarity and functionBiomaterials19961733733858745335
- WangYKYuXCohenDMBone morphogenetic protein-2-induced signaling and osteogenesis is regulated by cell shape, RhoA/ROCK, and cytoskeletal tensionStem Cells Dev20122171176118621967638
- LaiCFChengSLAlphavbeta integrins play an essential role in BMP-2 induction of osteoblast differentiationJ Bone Miner Res200520233034015647827
- DuJChenXLiangXIntegrin activation and internalization on soft ECM as a mechanism of induction of stem cell differentiation by ECM elasticityProc Natl Acad Sci U S A2011108239466947121593411
- PantovicAKrsticAJanjetovicKCoordinated time-dependent modulation of AMPK/Akt/mTOR signaling and autophagy controls osteogenic differentiation of human mesenchymal stem cellsBone201352152453123111315
- BjornstiMAHoughtonPJThe TOR pathway: a target for cancer therapyNat Rev Cancer20044533534815122205
- YehLCMaXFordJJAdamoMLLeeJCRapamycin inhibits BMP-7-induced osteogenic and lipogenic marker expressions in fetal rat calvarial cellsJ Cell Biochem201311481760177123444145
- LeeDYLiYSChangSFOscillatory flow-induced proliferation of osteoblast-like cells is mediated by alphavbeta3 and beta1 integrins through synergistic interactions of focal adhesion kinase and Shc with phosphatidylinositol 3-kinase and the Akt/mTOR/p70S6K pathwayJ Biol Chem20102851304219889638
- OgawaTTokudaMTomizawaKOsteoblastic differentiation is enhanced by rapamycin in rat osteoblast-like osteosarcoma (ROS 17/2.8) cellsBiochem Biophys Res Commun199824912262309705862
- SinghaUKJiangYYuSRapamycin inhibits osteoblast proliferation and differentiation in MC3T3-E1 cells and primary mouse bone marrow stromal cellsJ Cell Biochem2008103243444617516572
- VinalsFLopez-RoviraTRosaJLVenturaFInhibition of PI3K/p70 S6K and p38 MAPK cascades increases osteoblastic differentiation induced by BMP-2FEBS Lett20025101–29910411755539
