Abstract
Purpose
X-Ray Repair Cross Complementing 1 (XRCC1) functioning in the base excision repair pathway plays an important role in the repair of DNA single-strand breaks caused by ionizing radiation. The relationship between XRCC1 polymorphisms and the risk of radiation-induced side effects on normal tissues remains controversial. Therefore, we performed a comprehensive meta-analysis to elucidate these associations.
Materials and methods
A systematic literature search was carried out in PubMed, Medline (Ovid), Embase, Web of Science, Cochrane database, and the references of relevant studies. The pooled odds ratios (ORs) with corresponding 95% confidence intervals (CIs) were calculated to evaluate the strength of the association.
Results
A total of 40 studies including 6,682 patients were eventually identified in this meta-analysis. Pooled results suggested that rs25487 Arg399Gln polymorphism significantly increased the risk of acute radiation-induced side effects (OR=1.29, 95% CI: 1.10–1.52, P=0.002), especially acute mucositis (OR=1.91, 95% CI: 1.17–3.11, P=0.01) and acute gastrointestinal and genitourinary toxicity (OR=1.49, 95% CI: 1.04–2.11, P=0.03). Furthermore, patients who received head and neck irradiation with rs25487 Arg399Gln polymorphism were more likely to experience radiotherapy (RT)-induced side effects (OR=1.46, 95% CI: 1.12–1.90, P=0.005). However, no statistically significant correlations were identified between rs25487 polymorphism and any late side effects and other irradiation areas. Likewise, no significant associations were detected between rs25489, rs1799782, or rs3213245 polymorphism and RT-induced toxicity.
Conclusion
Our meta-analysis demonstrated that XRCC1 rs25487 Arg399Gln polymorphism had a significant predictive value and might predict a risk of severely acute RT-induced adverse effects, especially in acute mucositis and acute gastrointestinal and genitourinary toxicity, or in patients with head and neck irradiation. However, large-scale and well-designed studies are required to further evaluate the predictive value of XRCC1 variations on radiation-induced side effects in order to identify radiosensitive patients and predict radiotoxicity.
Keywords:
Introduction
Radiotherapy (RT) is a common and indispensable method in cancer treatment, which may result in a spectrum of normal tissue side effects.Citation1 Although improvements in precise RT techniques such as three-dimensional conformal radiotherapy and intensity-modulated radiotherapy have increased the possibility of dose escalation in tumor targets,Citation2 the implementation of radiation dose is still limited by the tolerance of normal tissues in and adjacent to the irradiation field.Citation3 However, patients exhibit substantially different degrees of normal tissue toxicity even with the same treatment regimen, varying from mild to severe and occasionally lethal. Acute adverse effects may lead to unanticipated RT breaks and then remarkably affect adequate treatment delivery,Citation4,Citation5 and late adverse effects markedly influence patients’ quality of life.Citation6 It is important to predict a predisposition of severe RT-induced adverse effects in normal tissues for making personal and optimized treatment decision, particularly in those with “high–intermediate risk”.
The severity of RT-induced complication is associated with many factors including irradiated dose, volume of normal tissues, fractionation schedule, combined with chemotherapy, as shown by Stone et al,Citation7 but they cannot fully explain patient-to-patient differences. Recent studies indicate that genetic component may contribute to the clinical radiosensitivity and radiation adverse effects.Citation8–Citation10 DNA is considered to be the main target of RT, which causes cell death by inducing base damage, single-strand breaks (SSBs), and double-strand breaks.Citation11 So, inter-individual differences in DNA repair capacity may determine varying degrees of the normal tissue response. Extensive researches have been conducted in order to identify some genetic markers such as single nucleotide polymorphisms (SNPs) as predictive factors for the risk of radiation-induced normal tissue toxicity.Citation10 SNPs in DNA damage and repair genes may alter the amino acid composition of encoded proteins, which play a role in individual’s radiation response and capacity of DNA damage repair. The protein encoded by X-Ray Repair Cross Complementing 1 (XRCC1) gene, which functions in the base excision repair (BER) pathway, involves in the efficient repair of DNA SSBs caused by exposure to ionizing radiation.Citation12 The XRCC1 gene is mapped at human chromosome 19q13.2–13.3. The most common variants of XRCC1 gene are rs25487 Arg399Gln in exon 10, rs25489 Arg280His in exon 9, and rs1799782 Arg194Trp in exon 6.Citation13–Citation15
Although several studies have investigated the association of XRCC1 polymorphisms with clinically observed normal tissue adverse effects, the results are not consistent. It is not sufficient to form a reliable conclusion and consequently limit their clinical applicability as biomarkers. So, we performed a systematic review to investigate these associations. This is, to our knowledge, the first comprehensive meta-analysis of genetics studies on the association between XRCC1 polymorphisms and radiation-related adverse effects.
Materials and methods
Search strategy
A systematic literature search in PubMed, Medline (Ovid), Cochrane, Embase, Web of Science database, and the references of relevant articles was carried out to identify studies involving XRCC1 polymorphisms and the risk of radiation-related normal tissue adverse effects (last search was updated on June 1, 2017). The search terms used were as follows: “XRCC1 or X-Ray Repair Cross Complementing 1” in combination with “SNP or polymorphism or variant or variation or mutation or haplotype” and “radiotherapy or radiation or irradiation” and “side effect or adverse effect or complication or injury or toxicity or reaction or response or radiotoxicity or radiosensitivity or morbidity or normal tissue”. All the search terms were restricted to studies in human subjects and in English language.
Inclusion criteria
Studies included in the current meta-analysis met the following inclusion criteria: 1) evaluation of the association between XRCC1 SNPs and radiation-induced normal tissue adverse effects; 2) the design has to be a cohort study or case control study; 3) sufficient published data (genotype distributions of each groups) to estimate an odds ratio (OR) with 95% CI.
Exclusion criteria
Studies were excluded if one of the following existed: 1) data of genotype frequencies of each group were not reported and 2) case reports, reviews, editorials, and repeat studies. If there were more than one study published by the same authors based on the same populations, the one providing the most comprehensive information was included.
Data extraction
Two investigators collected the data independently in duplicate according to the inclusion criteria listed above using a standardized data extraction form. The following items were extracted from each study: first author, publication date, original country, ethnicity, cancer type, subtype of SNP in XRCC1, normal tissue toxicity, sample size, treatment, type of study, genotyping method, genotype number, and total number in cases and controls.
Statistical analysis
ORs and 95% CIs were used to assess the strength of association between genetic polymorphisms and the risk of RT-induced adverse effects. The pooled OR was calculated by a fixed-effects model or a random-effects model according to the heterogeneity. Heterogeneity among eligible studies was measured by χ2-based Q-test and I2 statistical test.Citation16 If Q-test P<0.1 and I2-value ≥50%,Citation16 the heterogeneity was considered statistically significant, and the assumption of homogeneity was deemed invalid and the pooled OR was calculated by random-effects model after exploring the cause of heterogeneity. Otherwise, the fixed-effects model was used. Findings of our meta-analysis are shown in forest plots. The two-tailed P<0.05 was considered statistically significant. To evaluate the tolerance of different normal tissues and the occurrence time of side effects, subgroup analysis was conducted by early or late adverse effect, special types of side effects, and irradiation area. Sensitivity analysis was performed to confirm the stability and reliability of the pooled results by excluding each study individually and recalculating the pooled ORs and 95% CIs. If the number of included studies were >10, the possible publication bias and the degree of asymmetry were examined by Begg’s funnel plot and Egger’s test.Citation17,Citation18 If publication bias existed, the “trim and fill” methodCitation19 was used to estimate the number of missing studies and to adjust the pooled result. Statistical analysis was performed using Revman 5.3 and STATA 14.0 software.
Results
Study characteristics
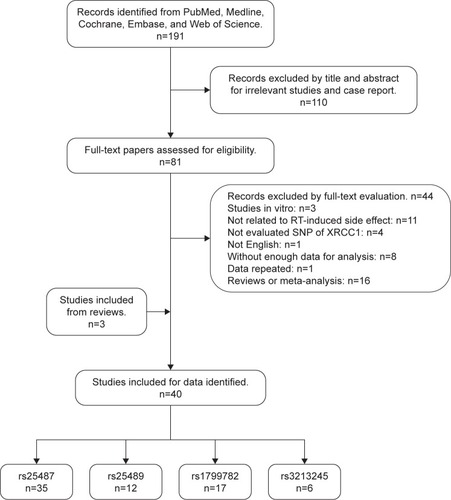
A total of 40 studies including 6,682 patients were eventually identified in this meta-analysis for further analysis (). The baseline characteristics of each included study are listed in . These studies were published from 2003 to 2017, and the sample size ranged from 34 to 579. Most of these studies included mainly Caucasian patients, and nine studies included Asian patients, of which five studies are on Chinese.Citation15,Citation20–Citation23 The cancer categories included head and neck cancer (8 studies),Citation20,Citation21,Citation23–Citation27 breast cancer (18 studies),Citation15,Citation28–Citation44 prostate cancer (5 studies),Citation14,Citation45–Citation48 cervical endometrial cancer (2 studies),Citation49,Citation50 bladder cancer (1 study),Citation51 rectal cancer (2 studies),Citation52,Citation53 non-small-cell lung cancer (NSCLC; 2 studies),Citation54,Citation55 esophageal cancer (2 studies),Citation22,Citation56 and one mixed cancers (mainly breast cancer and head and neck cancer).Citation57 Four subtypes of SNPs in XRCC1 were analyzed in this meta-analysis. Thirty-six studies were identified for rs25487, 12 studies for rs25489, 17 studies for rs1799782, and 6 studies for rs3213245. Subgroup analyses of radiation-induced adverse effects were performed on acute or late side effects, special types of side effects, and irradiation area. The genotype distributions analyzed in each study were in Hardy–Weinberg equilibrium with P>0.05. All the included eligible reports were written in English language.
Table 1 Baseline characteristics of the eligible studies
Figure 1 Flow diagram of study search and screening for the meta-analysis.
Meta-analysis results
XRCC1 rs25487 polymorphism
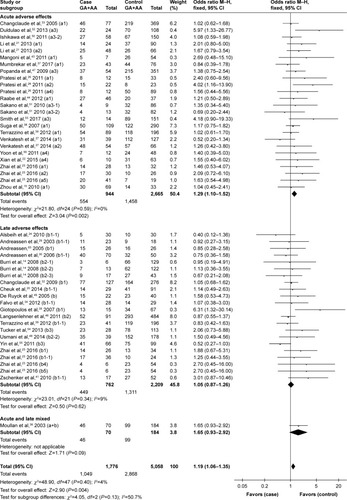
Overall, XRCC1 rs25487 Arg399Gln G>A polymorphism was significantly associated with acute normal tissue injury after RT. Specifically, rs25487 “Gln” allele increased the risk of acute radiation-induced adverse effects (for GA+AA versus GG, OR=1.29, 95% CI: 1.10–1.52, P=0.002; ).
Figure 2 Forest plot for the association between rs25487 and radiation-induced adverse effects.
Abbreviations: M–H, Mantel–Haenszel; OR, odds ratio.
Subgroup analysis by specific adverse effect
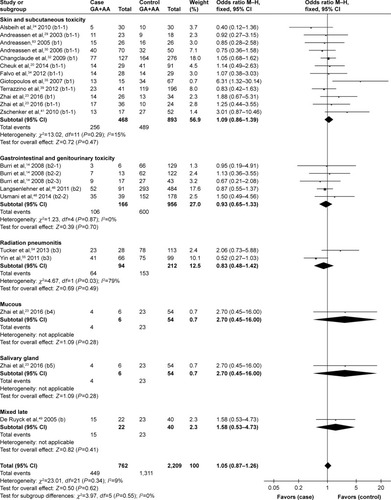
Because most studies investigated several different adverse effects, subgroup analysis was conducted by specific adverse effect. The results indicated that rs25487 Arg399Gln “Gln” allele carriers significantly increased acute mucositis (OR=1.91, 95% CI: 1.17–3.11, P=0.01) and acute gastrointestinal and genitourinary toxicity (OR=1.49, 95% CI: 1.04–2.11, P=0.03; ). No statistically significant associations were identified between rs25487 polymorphism and any late radiation-induced adverse effects ().
Figure 3 Forest plot for the association between rs25487 and radiation-induced acute adverse effects by specific side effect.
Abbreviation: M–H, Mantel–Haenszel.
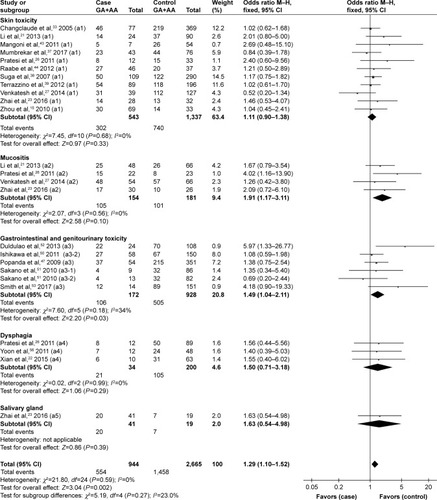
Figure 4 Forest plot for the association between rs25487 and radiation-induced late adverse effects by specific side effect.
Abbreviation: M–H, Mantel–Haenszel.
Subgroup analysis by radiotherapy area
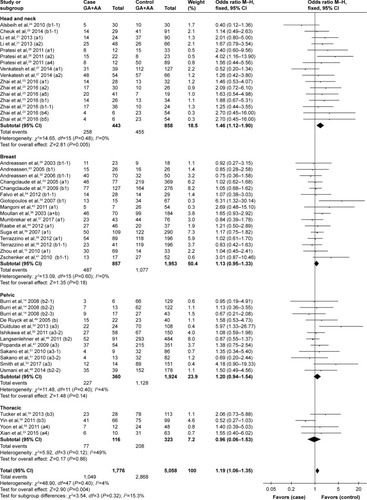
Subgroup analysis was conducted by different irradiation area irrespective of the type of adverse effect. The rs25487 Arg399Gln polymorphism was significantly associated with a higher risk of adverse effects induced by head and neck irradiation (OR=1.46, 95% CI: 1.12–1.90, P=0.005), whereas the correlation was not significant for breast or pelvic irradiation (breast, OR=1.13, 95% CI: 0.95–1.33, P=0.18; pelvic, OR=1.20, 95% CI: 0.94–1.54, P=0.14, respectively; ).
Figure 5 Forest plot for the association between rs25487 and radiation-induced adverse effects by irradiation area.
Abbreviation: M–H, Mantel–Haenszel.
XRCC1 rs25489, rs1799782, and rs3213245 polymorphisms
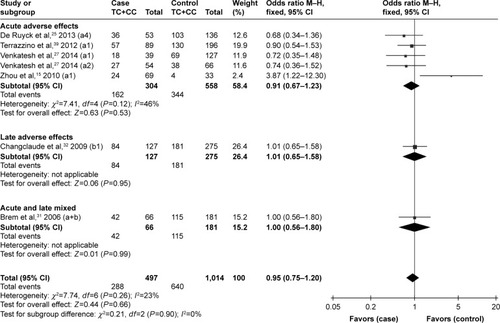
Although no statistically significant associations were identified, the rs25489 Arg280His polymorphism seemed to indicate a protective effect against radiotoxicity (OR=0.78, 95% CI: 0.58–1.06, P=0.11), especially in acute adverse effects (OR=0.66, 95% CI: 0.38–1.14, P=0.14, ) or in breast irradiation area (OR=0.71, 95% CI: 0.47–1.06, P=0.10, ). No significant associations were detected between rs1799782 or rs3213245 polymorphism and RT-induced toxicity ( and ).
Figure 6 Forest plot for the association between rs25489 and radiation-induced adverse effects.
Abbreviation: M–H, Mantel–Haenszel.
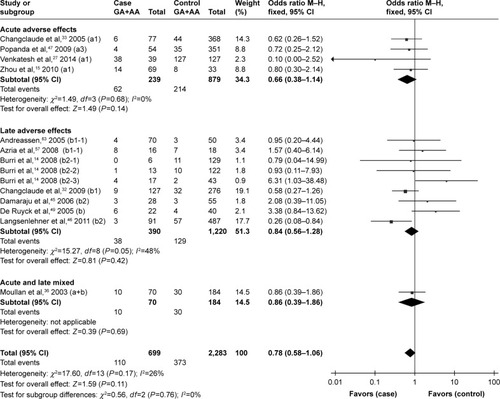
Figure 7 Forest plot for the association between rs25489 and radiation-induced adverse effects by irradiation area.
Abbreviation: M–H, Mantel–Haenszel.
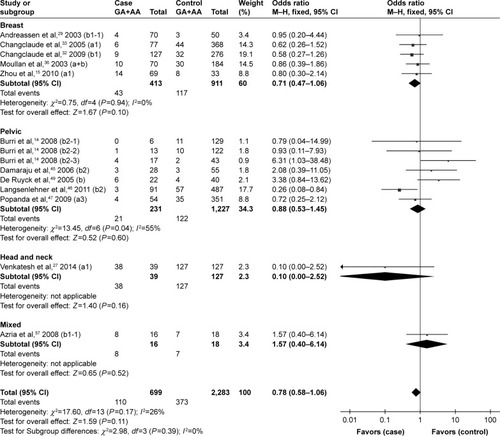
Figure 8 Forest plot for the association between rs1799782 and radiation-induced adverse effects.
Abbreviation: M–H, Mantel–Haenszel.
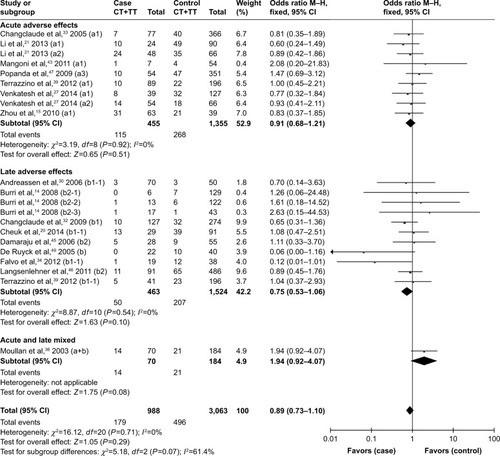
Figure 9 Forest plot for the association between rs3213245 and radiation-induced adverse effects.
Abbreviation: M–H, Mantel–Haenszel.
Heterogeneity and sensitivity analyses
The heterogeneities between studies of most analyses were not significant except for three subgroup analyses, the evaluation on radiation pneumonitis of rs25487 (I2=79%, χ2 P=0.03), late side effect (I2=48%, χ2 P=0.05), and pelvic irradiation (I2=55%, χ2 P=0.04) of rs25489. The pooled OR calculated by random-effects model of these subgroup analyses had no statistically significant associations, and the pooled results were stable in the sensitivity analysis.
Publication bias
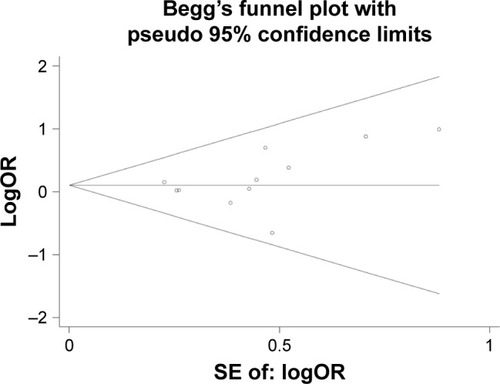
The distribution of all analyzed studies for rs25487 in Begg’s funnel plot was visually asymmetrical and the P-value of Egger’s test was significantly <0.05. However, we noticed that many included studies assessed several endpoints and different adverse effects, resulting in these studies being evaluated several times in Begg’s funnel plot, which led to an inaccurate result of the publication bias. So, in order to avoid “multiple testing problem”, we reevaluated the publication bias for each subgroup analysis if >10 studies were included based on the results above in the form of one study emerged only one time. The P-value of Egger’s test of rs25487 for RT-induced acute skin toxicity was 0.245 (), which indicates no publication bias. No publication bias was identified in other subgroup analysis of rs25487, and in rs25489, rs1799782, rs3213245 genetic models, and the P-values of Egger’s test were all >0.05, which suggested that there was no obvious risk of publication bias in the meta-analysis.
Discussion
The protein encoded by XRCC1 gene functions in the efficient repair of base damage and DNA SSBs formed by ionizing radiation and alkylating agents. This protein interacts with DNA ligase III, polymerase-beta, and poly (ADP-ribose) polymerase to participate in the BER pathway.Citation58 Polymorphisms in this gene are associated with varying radiosensitivity of cancer patients. Association studies on XRCC1 genetic variations and the risk of RT-induced normal tissue injuries can help us to identify markers predicting occurrence of side effects, but previous studies reported inconsistent findings. The present meta-analysis was performed to comprehensively evaluate the influence of XRCC1 polymorphisms on the development of radiation-induced normal tissue adverse effects. Four common SNPs of XRCC1 were analyzed in our meta-analysis: XRCC1 rs25487 (Arg399Gln, G>A), XRCC1 rs25489 (Arg280His, G>A), XRCC1 rs1799782 (Arg194Trp, C>T), and XRCC1 rs3213245 (−77 T>C). Among these, rs25487 (Arg399Gln, G>A) was the most commonly studied polymorphism of XRCC1 in previous researches. Due to different molecular mechanisms of acute and late radiation effects, we analyzed the acute and late side effects separately.
To date, several systematic reviews have been published on genetic variants and normal tissue toxicities induced by radiation, most of which involved XRCC1 polymorphism.Citation47,Citation59–Citation63 However, due to obvious heterogeneity, it is difficult to draw any definite conclusion. So far, four meta-analyses have been published on XRCC1 polymorphism and the risk of normal tissue injury after RT, three of which were performed only in breast cancer and one in prostate cancer patients; besides, only one to three polymorphisms have been analyzed in each paper.Citation13,Citation46,Citation64,Citation65 A positive association between rs25487 Arg399Gln polymorphism and acute side effect in breast cancer patients,Citation64,Citation65 and a negative association between rs25489 Arg280His variant and late side effect in breast cancer and prostate cancer patientsCitation46,Citation65 have been reported in these meta-analyses.
In our meta-analysis, more specific evidences were provided. For rs25487 Arg399Gln polymorphism, significant associations with seriously acute adverse effects were revealed, especially acute mucositis and acute gastrointestinal and genitourinary toxicity. Subgroup analysis according to irradiated area revealed that rs25487 Arg399Gln significantly correlated with an elevated risk of side effects induced by head and neck irradiation. It indicates that patients with rs25487 variant who receive RT are more likely to experience acute adverse effects, especially in head and neck irradiation. No significant correlation with any late side effects, or with breast, pelvic, or thoracic irradiation, was observed in rs25487 polymorphism. For rs25489 Arg280His variant, inconsistent with previous results reported, no statistically significant associations were identified, but rs25489 seemed to indicate a protective effect against radiotoxicity, especially in acute adverse effects or in breast irradiation. XRCC1 SNPs appear to be more likely to correlate with acute RT-induced side effects, but the reason is unclear. Radiation causes DNA strand breaks in normal cells, most of the cells die and cannot renew in time leading to acute side effects, accompanied by responses of DNA damage repair. Late side effects refer to the cells unable to regenerate after exhausted by radiation and eventually lead to fibrosis instead. The XRCC1 protein functions in the efficient repair of DNA SSBs; thus, we speculate that XRCC1 may participate in the DNA damage repair mainly in the period of RT-induced acute reactions.
No significant associations were detected between rs1799782 or rs3213245 polymorphism and RT-induced toxicity in the overall or the subgroup analyses. However, Moullan et alCitation36 and Mangoni et alCitation43 indicated that the rs1799782194Trp variant was associated with an increased risk of RT-induced adverse response when analyzed in combination with the rs25487 399Gln variant in breast cancer patients. No definite conclusion can be made for rs25489, rs1799782, or rs3213245 polymorphisms, may be due to the relatively small number of identified studies.
Although a series of studies have been made to evaluate the association between SNPs and RT-induced adverse effects, no SNPs have been thoroughly identified to have the predictive power in clinical practice. Moreover, most studies assessed the individual effect of selected SNPs,Citation59,Citation61–Citation63 and the original researches available on combined effect of multiple SNPs are less and not enough to make a meta-analysis. Further studies are needed to elucidate the selection criteria and predictive effect for SNP combinations. In addition, genome-wide association study (GWAS) is more credible due to the comprehensive genetic coverage. Barnett et alCitation66 presented the largest GWAS in which 1217 breast cancer patients received adjuvant RT and 633 prostate cancer patients received radical RT. Quantile–quantile plot results provided evidence for the true association between common genetic variants and late toxicity, and associations with late toxicity appeared to be tumor site-specific.
The main source of heterogeneity in such meta-analysis is the overall assessment of all kinds of side effects in various cancer types. However, there are two types of RT-induced adverse effects; acute side effects can be observed during RT and within several weeks after RT, while late side effects occur months to years later.Citation3 Furthermore, different RT-induced side effects may occur in the same irradiation area, while the same type of side effect can occur in different irradiation areas with the same histological structure. Hence, it is rational to make subgroup analysis in acute or late side effects, the special type of side effects, and irradiation areas.Citation67 The subgroup analyses evaluating the effect of rs25487 on radiation pneumonitis or on thoracic irradiation yielded significant heterogeneity, because the only two identified studies on NSCLC reached contrary conclusions. The evaluations of rs25489 on late side effects and pelvic irradiation as well as rs3213245 on acute side effects also yielded significant heterogeneity. The presence of heterogeneity may be caused by the differences in study characteristics such as treatment regimen, evaluation endpoint, and genotyping method. The results are reliable because the pooled results calculated by random-effects model are stable in the sensitivity analysis.
Limitations
Several limitations of the present meta-analysis should be considered. First, many included studies assessed multiple different endpoints, resulting in the same study being evaluated more than one time in one analysis. The “multiple testing problem” reduced the statistical power. Second, the number of trails in some of the subgroups and the sample sizes of some of the studies were relatively small, which also restricted the statistical power. Third, eight studies without sufficient data could not be evaluated by weight in the pooled result, which may cause some potential bias. Furthermore, data analyses were not stratified by other confounding factors such as ethnicity, genotyping method, radiation dose, or chemotherapy status because of insufficient information from the primary publications.
Conclusion
The present study, to our knowledge, is the first comprehensive meta-analysis of genetics studies on the association between XRCC1 polymorphisms and radiation-related adverse effects. In conclusion, the meta-analysis suggests that rs25487 Arg399Gln polymorphism is significantly associated with the risk of acute RT-induced adverse effects such as acute mucositis and acute gastrointestinal and genitourinary toxicity. Patients who received head and neck irradiation with rs25487 Arg399Gln polymorphism were more likely to experience RT-induced side effects. The present study also indicates a radioprotective effect for rs25489 polymorphism, especially in acute side effects or in breast irradiation, but without statistical significance. Well-designed studies with large sample size are needed to be performed to assess the value of XRCC1 polymorphisms on radiation-induced adverse effects, which can be used clinically to identify radiosensitive patients and predict radiotoxicity.
Acknowledgments
This work was supported by the Traditional Chinese Medicine Administration of Hebei Province (grant no. 2012018).
Disclosure
The authors report no conflicts of interest in this work.
References
- ChenYTrottiAColemanCNAdverse event reporting and developments in radiation biology after normal tissue injury: International Atomic Energy Agency consultationInt J Radiat Oncol Biol Phys20066451442145116414207
- PignonJPle MaitreAMaillardEBourhisJGroup M-NCMeta-analysis of chemotherapy in head and neck cancer (MACH-NC): an update on 93 randomised trials and 17,346 patientsRadiother Oncol200992141419446902
- CitrinDCotrimAPHyodoFBaumBJKrishnaMCMitchellJBRadioprotectors and mitigators of radiation-induced normal tissue injuryOncologist201015436037120413641
- BentzenSMSaundersMIDischeSBondSJRadiotherapy-related early morbidity in head and neck cancer: quantitative clinical radiobiology as deduced from the CHART trialRadiother Oncol200160212313511439207
- GruberSDorrWTissue reactions to ionizing radiation-Oral mucosaMutat Res2016770Pt B29229827919336
- AndratschkeNMaurerJMollsMTrottKRLate radiation-induced heart disease after radiotherapy. Clinical importance, radiobiological mechanisms and strategies of preventionRadiother Oncol2011100216016620826032
- StoneHBColemanCNAnscherMSMcBrideWHEffects of radiation on normal tissue: consequences and mechanismsLancet Oncol20034952953612965273
- AndreassenCNSearching for genetic determinants of normal tissue radiosensitivity–are we on the right track?Radiother Oncol20109711820817285
- AndreassenCNDikomeyEParliamentMWestCMWill SNPs be useful predictors of normal tissue radiosensitivity in the future?Radiother Oncol2012105328328823245645
- BarnettGCColesCEElliottRMIndependent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis studyLancet Oncol2012131657722169268
- NakanoTXuXSalemAMHShoulkamyMIIdeHRadiation-induced DNA-protein cross-links: mechanisms and biological significanceFree Radic Biol Med201710713614527894771
- TomkinsonAESallmyrAStructure and function of the DNA ligases encoded by the mammalian LIG3 geneGene2013531215015724013086
- SeiboldPBehrensSSchmezerPXRCC1 polymorphism associated with late toxicity after radiation therapy in breast cancer patientsInt J Radiat Oncol Biol Phys20159251084109226072091
- BurriRJStockRGCesarettiJAAssociation of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancerRadiat Res20081701495918582155
- ZhouLXiaJLiHDaiJHuYAssociation of XRCC1 variants with acute skin reaction after radiotherapy in breast cancer patientsCancer Biother Radiopharm201025668121204762
- HigginsJPThompsonSGQuantifying heterogeneity in a meta-analysisStat Med200221111539155812111919
- BeggCBMazumdarMOperating characteristics of a rank correlation test for publication biasBiometrics1994504108811017786990
- EggerMDavey SmithGSchneiderMMinderCBias in meta-analysis detected by a simple, graphical testBMJ199731571096296349310563
- DuvalSTweedieRTrim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysisBiometrics200056245546310877304
- CheukIWYipSPKwongDLWuVWAssociation of XRCC1 and XRCC3 gene haplotypes with the development of radiation-induced fibrosis in patients with nasopharyngeal carcinomaMol Clin Oncol20142455324940494
- LiHYouYLinCXRCC1codon 399Gln polymorphism is associated with radiotherapy-induced acute dermatitis and mucositis in nasopharyngeal carcinoma patientsRadiat Oncol2013813123375119
- XianYHeXBaojianZXuZGeWDNA repair gene ERCC1 C118T polymorphism predicts sensitivity of recurrent esophageal cancer to radiochemotherapy in a Chinese populationThorac Cancer20156674174826557912
- ZhaiXMHuQCGuKWangJPZhangJNWuYWSignificance of XRCC1 Codon399 polymorphisms in Chinese patients with locally advanced nasopharyngeal carcinoma treated with radiation therapyAsia Pac J Clin Oncol2016121e125e13223910235
- AlsbeihGAl-HarbiNAl-HadyanKEl-SebaieMAl-RajhiNAssociation between normal tissue complications after radiotherapy and polymorphic variations in TGFB1 and XRCC1 genesRadiat Res2010173450551120334523
- De RuyckKDuprezFWerbrouckJA predictive model for dysphagia following IMRT for head and neck cancer: introduction of the EMLasso techniqueRadiother Oncol2013107329529923618501
- PratesiNMangoniMManciniIAssociation between single nucleotide polymorphisms in the XRCC1 and RAD51 genes and clinical radiosensitivity in head and neck cancerRadiother Oncol201199335636121704413
- VenkateshGHManjunathVBMumbrekarKDPolymorphisms in radio-responsive genes and its association with acute toxicity among head and neck cancer patientsPLoS One201493e8907924594932
- AndreassenCNAlsnerJOvergaardJTGFB1 polymorphisms are associated with risk of late normal tissue complications in the breast after radiotherapy for early breast cancerRadiother Oncol2005751182115878096
- AndreassenCNAlsnerJOvergaardMOvergaardJPrediction of normal tissue radiosensitivity from polymorphisms in candidate genesRadiother Oncol200369212713514643949
- AndreassenCNAlsnerJOvergaardMSorensenFBOvergaardJRisk of radiation-induced subcutaneous fibrosis in relation to single nucleotide polymorphisms in TGFB1, SOD2, XRCC1, XRCC3, APEX and ATM–a study based on DNA from formalin fixed paraffin embedded tissue samplesInt J Radiat Biol200682857758616966185
- BremRCoxDGChapotBThe XRCC1 -77T->C variant: haplotypes, breast cancer risk, response to radiotherapy and the cellular response to DNA damageCarcinogenesis200627122469247416829685
- ChangclaudeJAmbrosoneCBLillaCGenetic polymorphisms in DNA repair and damage response genes and late normal tissue complications of radiotherapy for breast cancerBr J Cancer200910010168019367277
- ChangclaudeJPopandaOKroppSAssociation between polymorphisms in the DNA repair genes, XRCC1, APE1, and XPD and Acute side effects of radiotherapy in breast cancer patientsClin Cancer Res200511134802480916000577
- FalvoEStrigariLCitroGSNPs in DNA repair or oxidative stress genes and late subcutaneous fibrosis in patients following single shot partial breast irradiationJ Exp Clin Cancer Res2012311722272830
- GiotopoulosGSymondsRPFowerakerKThe late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causesBr J Cancer20079661001100717325707
- MoullanNCoxDGAngeleSRomestaingPGerardJPHallJPolymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapyCancer Epidemiol Biomarkers Prev20031211 Pt 11168117414652276
- MumbrekarKDBola SadashivaSRKabekkoduSPGenetic variants in CD44 and MAT1A confer susceptibility to acute skin reaction in breast cancer patients undergoing radiation therapyInt J Radiat Oncol Biol Phys201797111827816361
- SugaTIshikawaAKohdaMHaplotype-based analysis of genes associated with risk of adverse skin reactions after radiotherapy in breast cancer patientsInt J Radiat Oncol Biol Phys200769368569317889263
- TerrazzinoSLaMPGambaroGCommon variants of GSTP1, GSTA1, and TGFβ1 are associated with the risk of radiation-induced fibrosis in breast cancer patientsInt J Radiat Oncol Biol Phys201283250451122079734
- TerrazzinoSMattinaPLMasiniLCommon variants of eNOS and XRCC1 genes may predict acute skin toxicity in breast cancer patients receiving radiotherapy after breast conserving surgeryRadiother Oncol2012103219920522248507
- ZschenkerORaabeABoeckelmannIKAssociation of single nucleotide polymorphisms in ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with clinical and cellular radiosensitivityRadiother Oncol2010971263220170971
- FalvoEStrigariLCitroGDose and polymorphic genes xrcc1, xrcc3, gst play a role in the risk of article developing erythema in breast cancer patients following single shot partial breast irradiation after conservative surgeryBMC Cancer20111129121749698
- MangoniMBisanziSCarozziFAssociation between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patientsInt J Radiat Oncol Biol Phys2011811525820708344
- RaabeADerdaKReutherSAssociation of single nucleotide polymorphisms in the genes ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with risk of severe erythema after breast conserving radiotherapyRadiat Oncol201276522537351
- DamarajuSMurrayDDufourJAssociation of DNA repair and steroid metabolism gene polymorphisms with clinical late toxicity in patients treated with conformal radiotherapy for prostate cancerClin Cancer Res20061282545255416638864
- LangsenlehnerTRennerWGergerAAssociation between single nucleotide polymorphisms in the gene for XRCC1 and radiation-induced late toxicity in prostate cancer patientsRadiother Oncol201198338739321345510
- PopandaOMarquardtJUChang-ClaudeJSchmezerPGenetic variation in normal tissue toxicity induced by ionizing radiationMutat Res20096671–2586919022265
- UsmaniNLeongNMartellKSingle-nucleotide polymorphisms studied for associations with urinary toxicity from (125)I prostate brachytherapy implantsBrachytherapy201413328529124656733
- De RuyckKVanEMClaesKRadiation-induced damage to normal tissues after radiotherapy in patients treated for gynecologic tumors: association with single nucleotide polymorphisms in XRCC1, XRCC3, and OGG1 genes and in vitro chromosomal radiosensitivity in lymphocytesInt J Radiat Oncol Biol Phys2005624114015990020
- IshikawaASugaTShojiYGenetic variants of NPAT-ATM and AURKA are associated with an early adverse reaction in the gastrointestinal tract of patients with cervical cancer treated with pelvic radiation therapyInt J Radiat Oncol Biol Phys20118141144115221050672
- SakanoSHinodaYSasakiMNucleotide excision repair gene polymorphisms may predict acute toxicity in patients treated with chemoradiotherapy for bladder cancerPharmacogenomics201011101377138721047201
- DuldulaoMPLeeWNelsonRAGene polymorphisms predict toxicity to neoadjuvant therapy in patients with rectal cancerCancer201311951106111223096768
- SmithJJWassermanIMilgromSASingle nucleotide polymorphism TGFβ1 R25P correlates with acute toxicity during neoadjuvant chemoradiotherapy in rectal cancer patientsInt J Radiat Oncol Biol Phys201797592428333014
- TuckerSLLiMXuTIncorporating single-nucleotide polymorphisms into the Lyman model to improve prediction of radiation pneumonitisInt J Radiat Oncol Biol Phys201385125125722541966
- YinMLiaoZLiuZFunctional polymorphisms of base excision repair genes XRCC1 and APEX1 predict risk of radiation pneumonitis in patients with non-small cell lung cancer treated with definitive radiation therapyInt J Radiat Oncol Biol Phys2011813e67e7321420246
- YoonHHCatalanoPGibsonMKGenetic variation in radiation and platinum pathways predicts severe acute radiation toxicity in patients with esophageal adenocarcinoma treated with cisplatin-based preoperative radiochemotherapy: results from the Eastern Cooperative Oncology GroupCancer Chemother Pharmacol201168486387021286719
- AzriaDOzsahinMKramarASingle nucleotide polymorphisms, apoptosis, and the development of severe late adverse effects after radiotherapyClin Cancer Res200814196284628818829510
- HortonJKWatsonMStefanickDFShaughnessyDTTaylorJAWilsonSHXRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaksCell Res2008181486318166976
- AndreassenCNAlsnerJGenetic variants and normal tissue toxicity after radiotherapy: a systematic reviewRadiother Oncol200992329930919683821
- RattayTTalbotCJFinding the genetic determinants of adverse reactions to radiotherapyClin Oncol (R Coll Radiol)201426530130824702740
- WestCMBarnettGCGenetics and genomics of radiotherapy toxicity: towards predictionGenome Med2011385221861849
- GhazaliNShawRJRogersSNRiskJMGenomic determinants of normal tissue toxicity after radiotherapy for head and neck malignancy: a systematic reviewOral Oncol201248111090110022939215
- AndreassenCNCan risk of radiotherapy-induced normal tissue complications be predicted from genetic profiles?Acta Oncol200544880181516332587
- ZhouYZhouWLiuQXRCC1 R399Q polymorphism and risk of normal tissue injury after radiotherapy in breast cancer patientsTumour Biol2014351212524292986
- XieXXOuyangSYJinHKWangHZhouJMHuBQPredictive value of XRCC1 gene polymorphisms for side effects in patients undergoing whole breast radiotherapy: a meta-analysisAsian Pac J Cancer Prev201213126121612823464416
- BarnettGCThompsonDFachalLA genome wide association study (GWAS) providing evidence of an association between common genetic variants and late radiotherapy toxicityRadiother Oncol2014111217818524785509
- AndreassenCNBarnettGCLangendijkJAConducting radiogenomic research–do not forget careful consideration of the clinical dataRadiother Oncol2012105333734023245646