Abstract
Photoperiodism, the ability to respond to seasonal varying day length with suitable life history changes, is a common trait in organisms that live in temperate regions. In most studied organisms, the circadian system appears to be the basis for photoperiodic time measurement. In insects this is still controversial: while some data indicate that the circadian system is causally involved in photoperiodism, others suggest that it may have a marginal or indirect role. Resonance experiments in the parasitic wasp Nasonia vitripennis have revealed a circadian component in photoperiodic time measurement compatible with a mechanism of internal coincidence where a two components oscillator system obtains information from dawn and dusk, respectively. The identity of this oscillator (or oscillators) is still unclear but possible candidates are the oscillating molecules of the auto-regulatory feedback loops in the heart of the circadian system. Here, we show for the first time the circadian oscillation of period and cryptochrome mRNAs in the heads of Nasonia females kept under short and long photoperiods. Period and cryptochrome mRNA levels display a synchronous oscillation in all conditions tested and persist, albeit with reduced amplitude, during the first day in constant light as well as constant darkness. More importantly, the signal for the period and cryptochrome oscillations is set by the light-on signal. These results, together with phylogenetic analyses, indicate that Nasonia’s period and cryptochrome display characteristics of homologous genes in other hymenopteran species.
Introduction
Organisms that live in temperate regions react to seasonal changes with specific adaptations such as migration, hibernation or diapause. The photophase, the length of the light portion in a light:dark (LD) cycle, is a robust signal that reliably correlates with seasonal changes and many organisms rely on changes in the photoperiod for their seasonal reactions (Bradshaw & Holzapfel, Citation2007; Oster et al., Citation2002; Saunders, Citation2009; Tan et al., Citation2004; Vaz Nunes & Saunders, Citation1999; Yanovsky & Kay, Citation2003). How these changes are detected, interpreted and translated into physiological adaptations is investigated in different organisms. It is commonly agreed that a photoperiodic response, the “overt expression of a photoperiodic phenotype as a function of static or changing day or night length” (Bradshaw & Holzapfel, Citation2010), can be divided into four events: (1) photoreception; (2) measuring night (or day) length and interpret them as long or short by a photoperiodic timer (or clock); (3) count and accumulate successive inductive cycles to some sort of internal threshold by a photoperiodic counter (Saunders, Citation1981); and (4) the transduction of information provided by above events to downstream events that determine seasonal responses like diapause (Goto, Citation2013; Koštál, Citation2011; Saunders, Citation2010; Saunders & Bertossa, Citation2011). Since the photoperiodic response is composed of discrete events, separate mechanisms may be expected for each one of them and these may vary among organisms (Koštál, Citation2011; Meuti & Denlinger, Citation2013; Saunders & Bertossa, Citation2011). Insects in particular show considerable variation in the cellular substrates involved in the circadian system (i.e. circadian clock) (Tomioka & Matsumoto, Citation2010), in the clock genes involved in the auto-regulatory feedback loop in the circadian clock (Rubin et al., Citation2006; Sandrelli et al., Citation2008; Yuan et al., Citation2007), in life-history stages involved in the photoperiodic response (Denlinger, Citation2002; Saunders & Bertossa, Citation2011), and in the nature of the photoperiodic timer (Saunders, Citation2011).
In insects, an open question remains whether the circadian system is the functional basis of the photoperiodic timer (Koštál, Citation2011). More specifically, whether clock genes – the core molecular effectors of the circadian clock – affect the photoperiodic timer as a whole (“modular pleiotropy”), that is, as “circadian clock”, or individually (“gene pleiotropy”) (Bradshaw & Holzapfel, Citation2010; Emerson et al., Citation2009). In fact, measuring the length of the night (or day) does not require a circadian system but may be simply accomplished by an interval timer (a.k.a. hourglass timer), as in the vetch aphid, Megoura viciae (Saunders, Citation2011). There is a suggestion that a similar function (i.e. of interval timer) could be taken over by timeless – or timeless in combination with cryptochrome in Drosophila (Tauber & Kyriacou, Citation2001) – although other data suggest it may play a significant role more downstream of the photoperiodic timer (Saunders & Bertossa, Citation2011). While some evidence has been gathered in favor of hourglass-like timers, under specific conditions, also a dampened oscillator may produce results similar to those of an interval timer (Saunders & Lewis, Citation1987), making the search for a circadian influence on photoperiodism more difficult.
Many experimental findings support the hypothesis, advanced initially by Bünning (Citation1936), that the circadian system is somehow involved in photoperiodism (Saunders, Citation2010; Saunders & Bertossa, Citation2011). In vertebrates, for instance, the release of melatonin, which transmits photoperiod information to the body could be ascribed a role as interval timer. However, melatonin release is clearly dependent on the circadian system (Oster et al., Citation2002). In insects the strongest evidence so far comes from RNAi experiments in the bean bug Riptortus pedestris. Ikeno et al. (Citation2010, Citation2011a,Citationb,Citationc) showed that knocking down clock genes period (per), cryptochrome (cry) and cycle (cyc) disrupted circadian regulation of the alternate deposition of lamellate and non-lamellate cuticle layers as well as diapause. However, per or cry RNAi produced only non-lamellate cuticle layers and induced ovarian development in diapause-inducing short photoperiods whereas RNAi of cyc produced only lamellate layers and blocked ovarian development in diapause-preventing long photoperiods. These results are consistent with the opposing functions these clock genes have in the circadian clock and, therefore, suggest that the latter is the causal basis of circadian as well as photoperiodic traits in R. pedestris.
The parasitic wasp Nasonia vitripennis (Werren et al., Citation2010) has been used as a model to study photoperiodism in insects (Saunders, Citation1965, Citation1966, Citation1975; Saunders et al., Citation1970). Maternal induction of diapause in Nasonia shows a sharp reaction to changes in both photoperiod and thermoperiod (Saunders, Citation1973) typical for photoperiodic traits. By means of resonance experiments, Saunders could demonstrate a clear connection between the circadian clock and photoperiod measurement supporting the internal coincidence model envisaged by Pittendrigh (Citation1972). As Saunders (Citation1974) wrote, “The ‘ascending’ and ‘descending’ slopes of the maxima of diapause induction revealed by the resonance experiments may be interpreted as two components obtaining their principal time cues from dusk and dawn, respectively. Since they recur with circadian frequency in the extended dark periods used in this type of experiment, they are regarded as oscillators rather than ‘hour glasses’ which would not recur in such a manner”. The nature of these oscillators has remained elusive ever since in Nasonia. More recently, Bertossa et al. (Citation2010, Citation2013) have showed that eclosion but not emergence follows a circadian pattern in Nasonia wasps, and that these display long internal rhythms in constant darkness but short in constant light as well as considerable sex- and species-specific variation. Intriguingly, the presence of bimodal activity patterns and rhythm splits observed under constant conditions in Nasonia is compatible with the dual oscillator hypothesis (Pittendrigh & Daan, Citation1976). Furthermore, a latitudinal gradient in diapause induction was also discovered, suggesting that diapause adaptation to local conditions was probably influenced by photoperiod in Nasonia wasps (Paolucci et al., Citation2013). In order to understand whether and how the circadian system is connected to photoperiodism in this species, an important piece of information is whether clock genes products oscillate and how they react to a changing photoperiod.
In this study, we have analyzed the sequence, phylogenetic relationship within the insects, and the circadian levels of period and cryptochrome mRNAs, two key players in the circadian clock, in the heads of N. vitripennis females. Wasps were kept in short 12 h dark–12 h light (LD12:12) and long (LD18:6) photoperiods, as well as in constant darkness and light following entrainment in the same photoperiods. Results are discussed in the light of a possible mechanism for photoperiod measurement in Nasonia.
Materials and methods
Nasonia strain and rearing
Nasonia vitripennis (Hymenoptera: Pteromalidae) is a parasitoid wasp that parasitizes Sarcophagidae, Muscidae and Calliphoridae fly pupae (Campbell et al., Citation1993). Nasonia wasps have haplo-diploid reproduction (females are diploid and males are haploid): mated females produce broods with females and males, whereas unmated females can produce only males. After having drilled a hole into the fly puparium, females inject venom into the fly pupa onto which eggs are subsequently laid. At 25 °C offspring hatch within 48 h and begin feeding from the fly pupa. Approximately eight days after oviposition the larvae pupate and six days later adults eclose from the pupal integument. Next, males chew an exit hole through the host puparium and emerge. In mixed broods, N. vitripennis females typically emerge after males have emerged, as they require somewhat longer developmental time.
The laboratory inbred strain N. vitripennis AsymC was used for all experiments. This wild-type line collected in the Netherlands was cured from Wolbachia infection and maintained in the lab since 1971 (van den Assem & Jachmann, 1999). Insect rearing proceeded as previously described (Bertossa et al., Citation2010). In short, wasps were kept in mass culture vials (70 × 20 mm) at 25 °C, under a light:dark 18 h:6 h (LD18:6) cycle and around 45% relative humidity. For lab maintenance, about 25–30 wasps (females with some males) were transferred to new vials containing about 50 Calliphora sp. fly pupae, on which N. vitripennis females parasitize. After 14 days (at 25 °C) the emerging progeny were rehosted on fresh pupae in a similar way. Calliphora flies (mostly C. vomitoriae) were obtained as last instar larvae from a commercial manufacturer (Kreikamp & zn, Hoevelaken, The Netherlands). After pupation at room temperature in the lab, fly pupae were stored at 4 °C and used within four weeks.
The experimental procedures used in this study conform to international ethical standards as expressed by Portaluppi et al. (Citation2010).
Specimen collection and light conditions
Mated N. vitripennis females were allowed to oviposit individually on Calliphora pupae at 25 °C, LD18:6, and offspring to develop at the same conditions. At the yellow pupal stage (approximately eight days after eggs were laid), female offspring were collected and put into mass culture tubes in groups of approximately 35 females per tube. Half of the tubes was put at LD12:12 (short photoperiod) and half remained at LD18:6 (long photoperiod). Nine days later, virgin females eclosed in the mass culture tubes and were provided with hosts (two hosts per female). Hosts were replaced every other day (i.e. on the eclosion day, as well as two and four days after eclosion, ).
Figure 1. Overview of experimental set up. From a common LD18:6 entraining phase at 25 °C, wasps were split into six different conditions nine days before eclosion. On day 5, post-eclosion wasps were collected. Fresh hosts were provided every two days starting from eclosion day. On top, the life stages are indicated. Light and black boxes represent light and dark phases of days.
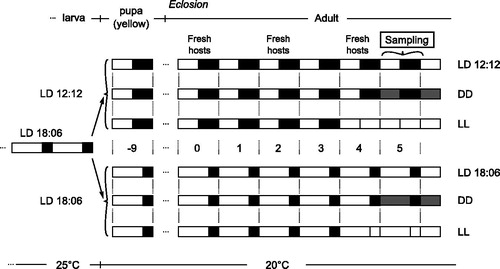
Four days after eclosion, some wasps from both short and long photoperiods were further shifted to constant light at the end of the light phase of day 4 (i.e. they did not enter the dark phase); the other wasps were kept at constant darkness beginning from the dark phase of day 4. Therefore, in total there were six light treatments: short and long photoperiods, constant light and constant darkness (after either short or long photoperiod).
Virgin wasps in mass culture tubes were collected on day 5 post-eclosion. Beginning from clock time 09:00 h (i.e. one hour after light-on), three mass culture tubes for each of the six light conditions were put into liquid nitrogen (to instantly kill the wasps) and stored immediately at −80 °C. For wasps reared in constant darkness this procedure was performed in constant darkness. Three new tubes with wasps were similarly frozen every 3 h until and included clock time 09:00 of the following day (day 6 post-eclosion) yielding nine consecutive samples of three tubes each. Wasps were later carefully separated from hosts while on liquid nitrogen and stored at −80 °C.
Heads collection, RNA extraction, cDNA synthesis, qPCR
Heads of wasps from one tube were collected and pooled as described previously (Bertossa et al., 2009). Total RNA was extracted from each heads pool with RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). 200 ng from each heads pool RNA sample were used to synthesize cDNA with Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Scientific, Waltham, MA). Before use in RT-qPCR, cDNA was diluted 10 times. Real-time qPCR was performed with the “qPCR GreenMaster with Rox” kit (Jena Bioscience, Jena, Germany). 2 µl of cDNA were used for each qPCR reaction. Two qPCR reactions per sample were run on an Applied Bioscience 7500 machine (Applied Biosystems, Foster City, CA). The qPCR profile was: 2′ at 95 °C for one cycle followed by 15′′ at 95 °C, 20′′ at 60 °C, and 30′′ at 72 °C for 40 cycles. Primers used in qPCR targeted the N. vitripennis genes period (NCBI Ref. Seq. XM_001604856.2, fwd.: ATCACGCTCTCTCGGGAAAT, rev.: TTCTTGGCTATGGTTGATGGTAT), cryptochrome (NCBI Ref. Seq. XM_001606355.2, fwd.: ACAAGAAGCAGCAGCAAAATTC, rev.: GATATTGATGTTGATGAACGTGTTG) and Elongation Factor 1α (EF-1a, NCBI Ref. Seq. NM_001172756.1, fwd.: CACTTGATCTACAAATGCGGTG, rev.: CCTTCAGTTTGTCCAAGACC). Expression data were first analyzed with LinRegPCR as described in Ramakers et al. (Citation2003) and then normalized to EF-1a, whose expression levels (not normalized) were constant throughout the period of analysis (data not shown). We refer to these data as “absolute” expression levels.
Expression data analysis and statistics
Analysis of expression levels was performed in R, version 2.13.0 (R Core Team, Citation2013). Gene expression data were divided by the mean expression level within each treatment and a sine wave model (gene expression ∼sin(time * (2 *pi/period) + phase) * amplitude + shift) was fitted to the data in order to determine circadian parameters. The nls2 function in R was used to determine the non-linear least-squares estimates of the model parameters (amplitude, phase, period, see ). The nls2 algorithm was “port”, maxiter was set to 1000, and parameters’ intervals were: phase = 0–360°, period = 10–35 h, amplitude = 0–3. The coefficient of determination R2 was calculated separately by computing the residual sum-of-squares (lack-of-fit) and the complement of its proportion to the total sum-of-squares. The probability (p) values () were obtained by randomizing the data and calculating the probability of obtaining the variance of the model’s residuals (“p randomized data”, ) as well as by comparing the sine wave model against a linear model with a Chi-squared test (“p sine wave versus Linear”, ). ANOVA was used to test the significance of the difference in expression levels among treatments.
Table 1. Period and cryptochrome sine wave fit parameters. Values represent the essential parameters (amplitude, period, phase, coefficient of determination R2 and p values) and 95% confidence intervals describing the sine wave fitting the oscillation of per and cry mRNAs data.
Phylogenetic and domain analyses
Protein sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov/) by searching for homologous genes in different species as well as performing cross BLAST searches to exclude the existence of possible paralogues or pseudogenes in the genomes. Protein sequences were aligned and evaluated with the Guidance algorithm (Penn et al., Citation2010b) on the respective server (Penn et al., Citation2010a) by applying 100 bootstrapping. Unreliable columns (below confidence score of 0.93) were removed. Amino acid substitution models were determined with MEGA5 (Tamura et al., Citation2011) using the Jones–Taylor–Thornton (JTT) for Period and rtREV for Cryptochrome. Gamma distributed across-site variation and four rate categories were used in both models. Bayesian analyses were performed with MrBayes 3.2.1 (Ronquist & Huelsenbeck, Citation2003) (http://mrbayes.sourceforge.net/index.php). Two parallel runs and four Monte Carlo Markov Chains searches per run were performed until the average standard deviation of split frequencies was less than 0.01 (500 000 generations for Period and 2 000 000 for Cryptochrome). Maximum Likelihood (ML) phylogenies were calculated in MEGA5 and support for tree topology obtained by bootstrap analysis (1000 replicates).
SMART (Letunic et al., Citation2012; Schultz et al., Citation1998) was used to scan the protein sequences for conserved domains. Phosphorylation sites were predicted with KinasePhos (Huang et al., Citation2005) at http://kinasephos.mbc.nctu.edu.tw. Nuclear Localization Sequences (NLS) were predicted with NLStradamus (Nguyen Ba et al., Citation2009) with cut-offs of 0.6 and 0.1 for Per and Cry, respectively.
Species and GeneBank accession numbers of the sequences can be found in Table S1.
Results
per and cry mRNA circadian expression
Virgin N. vitripennis AsymC females were raised at 25 °C in LD18:6 and then split into LD12:12 (short photoperiod) and LD18:6 (long photoperiod) at the yellow pupal stage (approximately eight days after egg deposition) according to the scheme in . Adults were collected on day 5 post-eclosion every 3 h beginning from 1 h after light-on in both photoperiods. Total RNA was extracted (and pooled) from the heads of 35 Nasonia wasps raised at the same conditions, converted into cDNA and relative levels of per and cry mRNA quantified (see “Materials and methods” section for details).
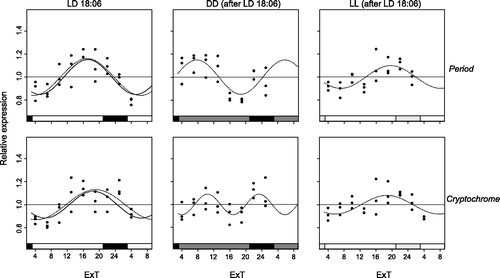
Both per and cry follow a circadian fluctuation in expression in LD as well as constant light conditions ( and ). However, oscillation amplitudes vary among them from a maximum of 0.26 for cry and 0.21 for per in LD to a minimum of 0.08 for cry and 0.1 for per in LL (after LD18:6, ). Oscillation amplitude is slightly smaller in the long photoperiod (LD18:6) than in the short one (LD12:12). However, this difference is only significant for cry, t(51.37) = 3.51, p = 0.00048, but not for per, t(44.80) = 1.39, p = 0.09 (compare and ). Oscillation of per and cry expression are synchronous in virtually all conditions. Even though the period of the oscillations varies across conditions (), the endogenous rhythm is usually greater than 24 h. The only exceptions are the period of per in LD12:12 when a free period is applied (23.6 h) as well as periods of both genes in DD (after LD18:6). Also for the absolute expression levels (i.e. clock gene transcript levels normalized to EF-1a transcript levels) there is a significant difference in the mean per transcript levels between LD12:12 (M = 0.18, SD = 0.03) and LD18:6 (M = 0.16, SD = 0.02), F(1, 49) = 6.69, p = 0.013, but no significant difference in average cry expression levels between LD12:12 (M = 0.017, SD = 0.003) and LD18:6 (M = 0.016, SD = 0.002), F(1, 49) = 0.47, p = 0.50.
Figure 2. Per and cry mRNA expression under LD12:12. Relative expression of period (top) and cryptochrome (bottom) mRNA in female Nasonia heads is indicated for every light condition: LD12:12 (left panels), as well as DD (middle) and LL (right) after an LD12:12 light regime. Each dot represents the expression value of a pool of 35 Nasonia heads. The curves represent the best sine wave fit of the experimental data for a fixed period of 24 h (continuous) or a free period (dashed line). Data are plotted according to External Time (Daan & Merrow, Citation2002). Filled bars on the bottom represent light (white and light gray) and dark (black and dark gray) phases. Light-on is at 6 h, light-off at 18 h ExT.
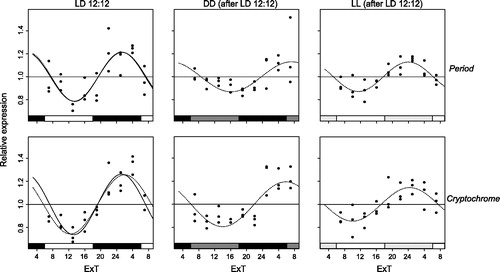
Figure 3. Per and cry mRNA expression under LD18:6. Relative expression of period (top) and cryptochrome (bottom) mRNA in female Nasonia heads is indicated for every light condition: LD18:6 (left panels), as well as DD (middle) and LL (right) after an LD18:6 light regime. Each dot represents the expression value of a pool of 35 Nasonia heads. The curves represent the best sine wave fit of the experimental data for a fixed period of 24 h (continuous) or a free period (dashed line). Data are plotted according to External Time (Daan & Merrow, Citation2002). Filled bars on the bottom represent light (white and light gray) and dark (black and dark gray) phases. Light-on is at 3 h, light-off at 21 h ExT.
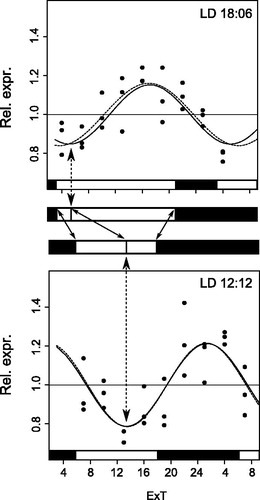
Changes in light conditions cause the phase of the oscillations to shift, the strongest being the change of photoperiod. Shift to a longer photoperiod causes the phase to be advanced. In fact, in LD12:12 the trough of the oscillation is in the middle/second half of the light phase whereas the peak is in the middle/second half of the dark phase (). In LD18:6 the trough is shifted towards the beginning of the light phase while expression peaks before light-off (). When changing from one to the other photoperiod the shift of the oscillation phase correlates with light-on (and not with light-off) and hence light-on is likely the signal which resets the oscillatory expression of per and cry mRNAs in the light-dark cycle ().
Figure 4. Per and cry mRNA expression tracks light-on. Scheme illustrating how both per and cry mRNA oscillations are set by the light-on signal: by changing photoperiod, the shift (dotted arrows) of any point in the sine wave (here the trough as example) correlates with the shift in the light-on instead of the light-off signal (continuous arrows in the “white-black” light scheme between both plots). Representative graphs are taken here from per. For other details see and .
Period and Cry phylogeny and their domains
Insect clock genes and their functional domains have been gained and lost in different insect lineages. This is reflected in different ways in which endogenous clocks can be built (Rubin et al., Citation2006; Sandrelli et al., Citation2008; Yuan et al., Citation2007). In order to understand their phylogenetic position, Nasonia Period and Cryptochrome were compared with homologous proteins in other taxa (, and S1). A distinct separation for the different Period homologues in insects is strongly supported. As expected, N. vitripennis Period clusters together with that of its hymenopteran relative, the honeybee Apis mellifera. The same is also true for Cryptochrome. Both species, together with the red flour beetle Tribolium castaneum, have lost insect Cryptochrome 1 which is still present in Dipterans and Lepidopterans, where it is part of the core transcriptional feed-back loop (Yuan et al., Citation2007).
Figure 5. Phylogenetic relationship of Period and Cryptochrome protein sequences. Period (A) and Cryptochrome (B) phylogenetic relationship with homologous proteins in other organisms were reconstructed with Maximum Likelihood (ML) and Bayesian methods. The Cryptochrome tree is rooted on a photolyase found in several taxa, including N. vitripennis. The Period tree is unrooted. Bayesian posterior probability (first) and ML bootstrap values (second) are separated by commas and indicated for relevant branches only when they are greater than 70%. N. vitripennis Cryptochrome is confusingly recorded as “Cryptochrome-1” in NCBI even though it is clearly of the vertebrate type (i.e. Cry2). N. vitripennis proteins are highlighted in bold. For amino acid substitution models see materials and methods. Species and GeneBank accession numbers for sequences used in the analyses can be found in Table S1 (supplementary material).
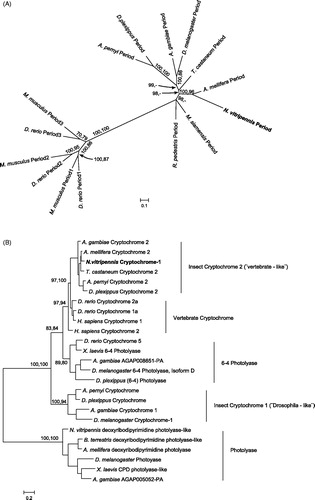
Figure 6. Putative functional domains in several Cryptochrome proteins. Functional domains in Cryptochrome proteins of several species are compared (A). Highlighted is the presence of domains important for vertebrate-like Cryptochromes (Hirayama & Sassone-Corsi, Citation2005) in Cry2 insect proteins, including that of N. vitripennis. Values underneath the domains are percentages of amino acid identity conservation relative to Homo sapiens Cry1 (on top). When identity percentage drops below 50% – as in Cry1 (“drosophila-like”) proteins – the domain is not filled (RD domains) or its contour is dashed (DNA photolyase). Known MAPK Serine phosphorylation sites relevant for vertebrate Crys function are indicated. Homologous Serine phosphorylation sites in insect Crys are indicated with a star. A detail of Cry proteins alignment around the central NLS (B) highlights its presence or absence in different species.
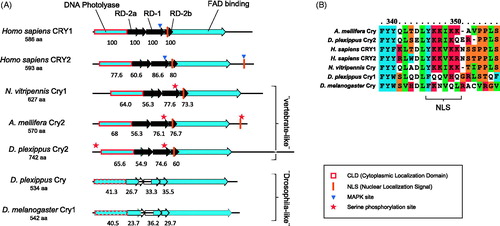
The picture emerging from the phylogenetic reconstruction is confirmed by an in-silico analysis of the most important domains contained in both proteins. Period in N. vitripennis contains domains present in all homologues, such as two PAS domains and the PAC domain which are needed for dimerization (Hirayama & Sassone-Corsi, Citation2005). However, only Nasonia Per and other vertebrate-like Per proteins contain the Cry-binding C-terminal domain (Figure S1). Nuclear localization signals (NLS) are consistently found in all insect Period proteins and may be homologous to those found in vertebrate Per proteins.
Insect Cryptochromes display an even greater diversity than Period proteins. In vertebrate-like Cryptochromes three domains are conserved and necessary for their repressive function: RD-1, RD-2a and RD-2b (Hirayama & Sassone-Corsi, Citation2005). CLD, a domain responsible for the cytoplasmic localization, is highly conserved in all vertebrate repressor-type Crys. Also in this case, vertebrate-like, as Nasonia Cry, but not drosophila-like Crys in insects show high similarity throughout all these domains (). A conserved NLS within the RD-2b region was found in the corresponding region in vertebrate-like Crys but not in Drosophila-like Crys. An additional NLS in the C-terminal region of vertebrate Cry proteins (i.e. Cry2) also mediates nuclear transport but was only found in the C-terminal region of A. mellifera. A potential Serine phosphorylation site in Nasonia and other vertebrate-like Crys is homologous to a similar site in vertebrate Crys RD-1 domains. Based on our analyses, Nasonia Period and Cryptochrome have features identical to those found in other vertebrate-like – in particular A. mellifera – Per and Cry proteins.
Discussion
For a better understanding of the molecular clock in Nasonia wasps and its role in photoperiod time measurement, we monitored the expression of per and cry mRNA in the heads of female wasps under different photoperiods and analyzed the protein sequences in a phylogenetic context. N. vitripennis per and cry mRNAs levels display a synchronous oscillation in virtually all light conditions analyzed. mRNA levels of both genes increase during or at the end of the light phase in LD18:6 and LD12:12, respectively, and reach a maximum at the end of the light phase in LD18:6 or the middle of the dark phase in LD12:12. This means that the phase is advanced in the long photoperiod compared to the short one and that circadian oscillation of both genes’ mRNA is phase-set to light-on (). Still, additional factors influence the phase of the oscillation. In fact, if no other influence is assumed, oscillation of both genes in DD and LL should appear similar, having both oscillations been reset at the same time point (i.e. light-on of day 4, ). However, in DD and LL, after LD12:12 for instance, phases appear slightly different. The model fits the best sine wave across the data, irrespective of its biological relevance. The fact that in DD after LD18:6 the sine wave fits of per and cry appear different is probably caused by variability in the data. Since in all other conditions per and cry fits appear very similar we expect that a resampling of the data would produce similar fits. We did not observe differences in overall cry expression levels between short and long photoperiods, as observed for instance by Bajgar et al. (Citation2013) in the gut of the linden bug Pyrrhocoris apterus, but a marginal significant difference in per expression levels.
Circadian levels of per and cry2 mRNA have been monitored in the head of several insect species and similar results have been obtained. Per mRNA oscillation typically displays a peak generally at light-off or the first part of the dark phase and a trough in the light phase, irrespective of the photoperiod. This has been observed in the hymenopterans A. mellifera (Rubin et al., Citation2006), A. cerana (Shimizu et al., Citation2001), and Solenopsis invicta (Ingram et al., Citation2012); in the dipterans Drosophila melanogaster (Qiu & Hardin, Citation1996), A. aegypti, Culex quinquefasciatus (Gentile et al., Citation2009), Protophormia terranovae (Muguruma et al., Citation2010), Sarcophaga crassipalpis (Goto & Denlinger, Citation2002; Kostál et al., Citation2009), Chymomyza costata (Kobelková et al., Citation2010), and Sarcophaga bullata (Goto et al., Citation2006); in the cricket Modicogryllus siamensis (Sakamoto et al., Citation2009), the cockroach Rhyparobia maderae (Werckenthin et al., Citation2012), and in the lepidopterans Bombyx mori (Iwai et al., Citation2006), Danaus plexippus (Zhu et al., Citation2008), and Spodoptera littoralis (Merlin et al., Citation2007). In species where cry2 (or “vertebrate-like” cry) is present, its oscillation matches that of per, as found in our study, with a trough in the light phase and a peak in the dark phase (Gentile et al., Citation2009; Ikeno et al., Citation2008; Ingram et al., Citation2012; Merlin et al., Citation2007; Rubin et al., Citation2006; Werckenthin et al., Citation2012; Yan et al., Citation2013; Zhu et al., Citation2008). Therefore, oscillation of per and cry2 in the heads of N. vitripennis females is similar to what has been found in other insect species.
In some of these species the phase of per mRNA oscillation was also shifted when the photoperiod was changed. Like in N. vitripennis, light-on seems to set the phase of per mRNA cycling in M. siamensis (Sakamoto et al., Citation2009). Yet, in D. melanogaster, C. costata and R. maderae per mRNA cycling is phase-set by light-off (Kobelková et al., Citation2010; Qiu & Hardin, Citation1996; Werckenthin et al., Citation2012) and a similar result is observed in B. mori (Iwai et al., Citation2006).
Both per and cry mRNAs oscillate, albeit with reduced amplitude, also during the first day in constant light conditions, as found in other insects (Rubin et al., Citation2006; Werckenthin et al., Citation2012; Zhu et al., Citation2008). A slightly dampened oscillation was also observed in the long photoperiod compared to the short one. While dampening of per oscillation in a long photoperiod is also seen in some insects species (Ikeno et al., Citation2008; Syrová et al., Citation2003), in R. maderae dampening has been observed in the short photoperiod (Werckenthin et al., Citation2012). In mammals, dampening of clock genes expression in LL is explained by a reduced coupling of individual clock neurons in the suprachiasmatic nucleus (SCN), seat of the master clock (Ohta et al., Citation2005). Instead, in D. melanogaster, the split in locomotor activity rhythms between a short and a long component in constant light conditions is associated with decoupling among clock neurons (Rieger et al., Citation2006; Yoshii et al., Citation2004).
Different clock models in insects have been proposed based on the characteristic of Cry proteins to repress Clock:Bmal(Cycle)-mediated transcription and to be degraded by light (Yuan et al., Citation2007; Zhu et al., Citation2005). At one end of the spectrum there is the Drosophila-like clockwork in which Cryptochrome has a light-resetting function and the repressive function in the auto-regulatory feedback loop is taken over by Timeless. At the other extreme we have the vertebrate-like system where Cryptochromes (in combination with Period proteins) instead of Timeless are translocated into the nucleus to repress Clock:Bmal1-mediated transcription (Hirayama & Sassone-Corsi, Citation2005). The ancestral state in insects appears to comprise both versions of Cry, i.e. the Drosophila- as well as the vertebrate-like (Cry1 and Cry2, respectively). It has thus far been found in the lepidopterans D. plexippus and A. pernyi as well as the dipterans A. gambiae and C. quinquefasciatus (Gentile et al., Citation2009; Yuan et al., Citation2007). Because Cry1 but not Cry2 has a light-entraining function in some insect clocks, the presence or absence of cryptochromes in insect genomes has been suggested to be potentially symptomatic of external versus internal coincidence (Saunders, Citation2012). The phylogenetic analysis and the presence of conserved domains in N. vitripennis Per and Cry protein sequences suggest that both genes have functions similar to those found in insect species with the same genes and domains, namely the hymenopteran A. mellifera. Per is expected to dimerize with Cryptochrome through its C-terminal Cryptochrome-binding domain and together to suppress their own Clock:Cycle-mediated transcription. This however requires further experimental confirmation.
Resonance experiments in Nasonia have demonstrated a circadian component in photoperiodism with a dual oscillator, one driven by light-on the other by light-off, as probable mechanistic effector (Saunders, Citation1974). More recent experiments in D. melanogaster have revealed the presence of oscillating clock genes expression in morning (M) and evening (E) brain clock neurons responsible for morning and evening locomotor activity peaks, respectively (Grima et al., Citation2004; Stoleru et al., Citation2004). M-cells have master clock function for circadian output in a short photoperiod (LD10:14) whereas E-cells take over the master role in a long photoperiod (LD14:10). The system allows circadian output adjustment to different photoperiods (Stoleru et al., Citation2007). However, another possibility is a molecular double oscillator. In 2001, relying on the knowledge available at that time, Daan et al. (Citation2001) put forward an updated version of the original double oscillator hypothesis (Pittendrigh & Daan, Citation1976) in which mouse mPer1 and mCry1 would be elements of the M whereas mPer2 and mCry2 of the E oscillator. More recent findings in flies and mice led Helfrich-Förster (Citation2009) to conclude that the E and M hypothesis, because of missing conclusive data in those species, would perhaps be more applicable to strongly photoperiodic species. In Nasonia, while the oscillation of per and cry mRNAs is reset by light-on, another oscillating gene(s) could be reset by light-off. A candidate could be cycle (cyc). In the other hymenopterans for which the molecular clockwork is known, A. mellifera (Rubin et al., Citation2006) and S. invicta, cyc mRNA cycling in heads has clearly another phase than per and cry in LD12:12 but its oscillation in a longer photoperiod has not been tested yet. Therefore, another important step in understanding photoperiod measurement in N. vitripennis is monitoring the circadian oscillation of other clock gene mRNAs, cycle in particular, in short and long photoperiods.
Declaration of interest
RCB was supported by a grant (ALW 817.02.020) from the Netherlands Organization for Scientific Research (NWO) URL: http://www.nwo.nl/. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Open Access publication is supported by an “Incentive Fund Open Access Publications” grant from NWO.
The authors declare no conflicts of interest.
Supplementary Material
Download PDF (167.3 KB)Acknowledgments
We are grateful to Roelof Hut and Gerard te Meerman for statistical advice. We thank David Saunders and two anonymous reviewers for their valuable comments on a previous version of the manuscript.
References
- Bajgar A, Jindra M, Dolezel D. (2013). Autonomous regulation of the insect gut by circadian genes acting downstream of juvenile hormone signaling. Proc Natl Acad Sci USA. 110:4416–21
- Bertossa RC, van de Zande L, Beukeboom LW. (2009). The fruitless gene in Nasonia displays complex sex-specific splicing and contains new zinc-finger domains. Mol Biol Evol. 26:1557–69
- Bertossa RC, van Dijk J, Beersma DG, Beukeboom LW. (2010). Circadian rhythms of adult emergence and activity but not eclosion in males of the parasitic wasp Nasonia vitripennis. J Insect Physiol. 56:805–12
- Bertossa RC, van Dijk J, Diao W, et al. (2013). Circadian rhythms differ between sexes and closely related species of Nasonia wasps. PLoS One. 8:e60167
- Bradshaw WE, Holzapfel CM. (2007). Evolution of animal photoperiodism. Annu Rev Ecol Evol S. 38:1–25
- Bradshaw WE, Holzapfel CM. (2010). What season is it anyway? Circadian tracking vs. photoperiodic anticipation in insects. J Biol Rhythm. 25:155–65
- Bünning E. (1936). Die endonome Tagesrhythmik als Grundlage der photoperiodischen Reaktion. Ber Dtsch Bot Ges. 54:590–607
- Campbell BC, Steffen-Campbell JD, Werren JH. (1993). Phylogeny of the Nasonia species complex (Hymenoptera: Pteromalidae) inferred from an internal transcribed spacer (ITS2) and 28S rDNA sequences. Insect Mol Biol. 2:225–37
- Daan S, Albrecht U, Van der Horst GTJ, et al. (2001). Assembling a clock for all seasons: Are there M and E oscillators in the genes? J Biol Rhythm. 16:105–16
- Daan S, Merrow M. (2002). External time-internal time. J Biol Rhythm. 17:107–9
- Denlinger DL. (2002). Regulation of diapause. Annu Rev Entomol. 47:93–122
- Emerson KJ, Bradshaw WE, Holzapfel CM. (2009). Complications of complexity: Integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 25:217–25
- Gentile C, Rivas GBS, Meireles-Filho ACA, et al. (2009). Circadian expression of clock genes in two mosquito disease vectors: Cry2 is different. J Biol Rhythm. 24:444–51
- Goto SG. (2013). Roles of circadian clock genes in insect photoperiodism. Entomol Sci. 16:1–16
- Goto SG, Denlinger DL. (2002). Short-day and long-day expression patterns of genes involved in the flesh fly clock mechanism: Period, timeless, cycle and cryptochrome. J Insect Physiol. 48:803–16
- Goto SG, Han B, Denlinger DL. (2006). A nondiapausing variant of the flesh fly, Sarcophaga bullata, that shows arrhythmic adult eclosion and elevated expression of two circadian clock genes, period and timeless. J Insect Physiol. 52:1213–18
- Grima B, Chélot E, Xia R, Rouyer F. (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 431:869–73
- Helfrich-Förster C. (2009). Does the morning and evening oscillator model fit better for flies or mice? J Biol Rhythm. 24:259–70
- Hirayama J, Sassone-Corsi P. (2005). Structural and functional features of transcription factors controlling the circadian clock. Curr Opin Genet Dev. 15:548–56
- Huang H-D, Lee T-Y, Tzeng S-W, Horng J-T. (2005). KinasePhos: A web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 33:W226–9
- Ikeno T, Katagiri C, Numata H, Goto SG. (2011a). Causal involvement of mammalian-type cryptochrome in the circadian cuticle deposition rhythm in the bean bug Riptortus pedestris. Insect Mol Biol. 20:409–15
- Ikeno T, Numata H, Goto SG. (2011b). Photoperiodic response requires mammalian-type cryptochrome in the bean bug Riptortus pedestris. Biophs Res Commun. 410:394–7
- Ikeno T, Numata H, Goto SG. (2011c). Circadian clock genes period and cycle regulate photoperiodic diapause in the bean bug Riptortus pedestris males. J Insect Physiol. 57:935–8
- Ikeno T, Numata H, Goto SG. (2008). Molecular characterization of the circadian clock genes in the bean bug, Riptortus pedestris, and their expression patterns under long- and short-day conditions. Gene. 419:56–61
- Ikeno T, Tanaka SI, Numata H, Goto SG. (2010). Photoperiodic diapause under the control of circadian clock genes in an insect. BMC Biol. 8:116
- Ingram KK, Kutowoi A, Wurm Y, et al. (2012). The molecular clockwork of the fire ant Solenopsis invicta. PloS One. 7:e45715
- Iwai S, Fukui Y, Fujiwara Y, Takeda M. (2006). Structure and expressions of two circadian clock genes, period and timeless in the commercial silkmoth, Bombyx mori. J Insect Physiol. 52:625–37
- Kobelková A, Bajgar A, Dolezel D. (2010). Functional molecular analysis of a circadian clock gene timeless promoter from the drosophilid fly Chymomyza costata. J Biol Rhythm. 25:399–409
- Koštál V. (2011). Insect photoperiodic calendar and circadian clock: Independence, cooperation, or unity? J Insect Physiol. 57:538–56
- Kostál V, Závodská R, Denlinger D. (2009). Clock genes period and timeless are rhythmically expressed in brains of newly hatched, photosensitive larvae of the fly, Sarcophaga crassipalpis. J Insect Physiol. 55:408–14
- Letunic I, Doerks T, Bork P. (2012). SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40:D302–5
- Merlin C, Lucas P, Rochat D, et al. (2007). An antennal circadian clock and circadian rhythms in peripheral pheromone reception in the moth Spodoptera littoralis. J Biol Rhythm. 22:502–14
- Meuti ME, Denlinger DL. (2013). Evolutionary links between circadian clocks and photoperiodic diapause in insects. Integr Comp Biol 53:131–43
- Muguruma F, Goto SG, Numata H, Shiga S. (2010). Effect of photoperiod on clock gene expression and subcellular distribution of PERIOD in the circadian clock neurons of the blow fly Protophormia terraenovae. Cell Tissue Res. 340:497–507
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. (2009). NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics. 10:202
- Ohta H, Yamazaki S, McMahon DG. (2005). Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 8:267–9
- Oster H, Maronde E, Albrecht U. (2002). The circadian clock as a molecular calendar. J Biol Rhythms. 19:507–16
- Paolucci S, van de Zande L, Beukeboom LW. (2013). Adaptive latitudinal cline of photoperiodic diapause induction in the parasitoid Nasonia vitripennis in Europe. J Evol Biol. 26:705–18
- Penn O, Privman E, Ashkenazy H, et al. (2010a). GUIDANCE: A web server for assessing alignment confidence scores. Nucleic Acids Res. 38:W23–8
- Penn O, Privman E, Landan G, et al. (2010b). An alignment confidence score capturing robustness to guide tree uncertainty. Mol Biol Evol. 27:1759–67
- Pittendrigh CS. (1972). Circadian surfaces and the diversity of possible roles of circadian organization in photoperiodic induction. Proc Natl Acad Sci USA. 69:2734–7
- Pittendrigh CS, Daan S. (1976). A functional analysis of circadian pacemakers in nocturnal rodents. 5. Pacemaker structure: A clock for all seasons. J Comp Physiol A. 106:333–55
- Portaluppi F, Smolensky MH, Touitou Y. (2010). Ethics and methods for biological rhythm research on animals and human beings. Chronobiol Int. 27:1911–29
- Qiu J, Hardin PE. (1996). per mRNA cycling is locked to lights-off under photoperiodic conditions that support circadian feedback loop function. Mol Cell Biol. 16:4182–8
- R Core Team. (2013). A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available from: http://www.R-project.org
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 339:62–6
- Rieger D, Shafer OT, Tomioka K, Helfrich-Förster C. (2006). Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci. 9:2531–43
- Ronquist F, Huelsenbeck JP. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–74
- Rubin EB, Shemesh Y, Cohen M, et al. (2006). Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16:1352–65
- Sakamoto T, Uryu O, Tomioka K. (2009). The clock gene period plays an essential role in photoperiodic control of nymphal development in the cricket Modicogryllus siamensis. J Biol Rhythm. 24:379–90
- Sandrelli F, Costa R, Kyriacou CP, Rosato E. (2008). Comparative analysis of circadian clock genes in insects. Insect Mol Biol. 17:447–63
- Saunders DS. (1965). Larval diapause induced by maternally-operating photoperiod. Nature. 206:739–40
- Saunders DS. (1966). Larval diapause of maternal origin – II. The effect of photoperiod and temperature on Nasonia vitripennis. J Insect Physiol. 12:569–81
- Saunders DS. (1973). Thermoperiodic control of diapause in an insect: Theory of internal coincidence. Science. 181:358–60
- Saunders DS. (1974). Evidence for “dawn” and “dusk” oscillators in the Nasonia photoperiodic clock. J Insect Physiol. 20:77–88
- Saunders DS. (1975). Spectral sensitivity and intensity thresholds in Nasonia photoperiodic clock. Nature. 253:732–4
- Saunders DS. (1981). Insect photoperiodism – the clock and the counter: A review. Physiol Entomol. 6:99–116
- Saunders DS. (2009). Circadian rhythms and the evolution of photoperiodic timing in insects. Physiol Entomol. 34:301–8
- Saunders DS. (2010). Controversial aspects of photoperiodism in insects and mites. J Insect Physiol. 56:1491–502
- Saunders DS. (2011). Unity and diversity in the insect photoperiodic mechanism. Entomol Sci. 14:235–44
- Saunders DS. (2012). Insect photoperiodism: Seeing the light. Physiol Entomol. 37:207–18
- Saunders DS, Bertossa RC. (2011). Deciphering time measurement: The role of circadian “clock” genes and formal experimentation in insect photoperiodism. J Insect Physiol. 57:557–66
- Saunders DS, Lewis RD. (1987). A damped circadian oscillator model of an insect photoperiodic clock. J Theor Biol. 128:61–71
- Saunders DS, Sutton D, Jarvis RA. (1970). The effect of host species on diapause induction in Nasonia vitripennis. J Insect Physiol. 16:405–16
- Schultz J, Milpetz F, Bork P, Ponting CP. (1998). Colloquium Paper: SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA. 95:5857–64
- Shimizu I, Kawai Y, Taniguchi M, Aoki S. (2001). Circadian rhythm and cDNA cloning of the clock gene period in the honeybee Apis cerana japonica. Zool Sci. 18:779–89
- Stoleru D, Nawathean P, Mde LF, et al. (2007). The Drosophila circadian network is a seasonal timer. Cell. 129:207–19
- Stoleru D, Peng Y, Agosto J, Rosbash M. (2004). Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 431:862–8
- Syrová Z, Dolezel D, Saumann I, Hodková M. (2003). Photoperiodic regulation of diapause in linden bugs: Are period and clock genes involved? Cell Mol Life Sci. 60:2510–15
- Tamura K, Peterson D, Peterson N, et al. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–9
- Tan Y, Merrow M, Roenneberg T. (2004). Photoperiodism in Neurospora crassa. J Biol Rhythm. 19:135–43
- Tauber E, Kyriacou BP. (2001). Insect photoperiodism and circadian clocks: Models and mechanisms. J Biol Rhythms. 16:381–90
- Tomioka K, Matsumoto A. (2010). A comparative view of insect circadian clock systems. Cell Mol Life Sci. 67:1397–406
- Van den Assem J, Jachmann F. (1999). Changes in male perseverance in courtship and female readiness to mate in a strain of the parasitic wasp Nasonia vitripennis over a period of 20+ years. Neth J Zool. 49:125–37
- Vaz Nunes M, Saunders D. (1999). Photoperiodic time measurement in insects: A review of clock models. J Biol Rhythms. 14:84–104
- Werckenthin A, Derst C, Stengl M. (2012). Sequence and expression of per, tim1, and cry2 genes in the Madeira cockroach Rhyparobia maderae. J Biol Rhythm. 27:453–66
- Werren JH, Richards S, Desjardins CA, et al. (2010). Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 327:343–8
- Yan S, Ni H, Li H, et al. (2013). Molecular cloning, characterization, and mRNA expression of two cryptochrome genes in Helicoverpa armigera (Lepidoptera: Noctuidae). J Econ Entomol. 106:450–62
- Yanovsky MJ, Kay SA. (2003). Living by the calendar: How plants know when to flower. Nat Rev Mol Cell Biol. 4:265–75
- Yoshii T, Funada Y, Ibuki-Ishibashi T, et al. (2004). Drosophila cryb mutation reveals two circadian clocks that drive locomotor rhythm and have different responsiveeness to light. J Insect Physiol. 50:479–88
- Yuan Q, Metterville D, Briscoe AD, Reppert SM. (2007). Insect cryptochromes: Gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 24:948–55
- Zhu H, Sauman I, Yuan Q, et al. (2008). Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 6:e4
- Zhu H, Yuan Q, Froy O, et al. (2005). The two CRYs of the butterfly. Curr Biol. 15:R953–4

