Abstract
Background and aim: Streptococci are a heterogeneous group that includes more than 30 different species. Most of the species are usual components of the human flora and some are important human pathogens. To date, molecular tools have not been developed for studying the diversity of the genus Streptococcus in clinical samples. We have developed a PCR-based analytical tool to investigate the presence of new yet uncultured pathogenic streptococci in clinical samples. Methods: A 16S rRNA gene-targeted oligonucleotide (1043F) covering all species of the genus Streptococcus has been designed and used in combination with the universal eubacterial primer, 1492R, to produce approximately 450 bp PCR fragments that enclose three (V7, V8 and V9) hypervariable regions. PCR products are suitable for separation through denaturing gradient gel electrophoresis (DGGE). The inclusivity of the PCR assay was verified on 16 different Streptococcus species representing all current taxonomic groups. The exclusivity of the primers was tested on 29 other bacterial species representing all major phylogenetic lineages. Results: Positive PCR results were obtained for all Streptococcus species and Enterococcus faecalis, whereas PCR was negative for the other specimens. The amplification limit was 170 genome equivalents per reaction (95% probability). The oligonucleotide set was tested on colon biopsies and bronchoalveolar lavage samples. Thus, from the 33 sequences retrieved, 16 (48.5%) shared identity to cultured streptococci, whereas 11 (33.3%) of the sequences belonged to uncultured streptococci and three (9%) to other genera. Three sequences (9.7%) corresponded to new Streptococcus phylotypes (< 95% identified to previously known sequences). Conclusion: The use of this new primer set in combination with DGGE has proven to be a suitable method for studying the specific composition of streptococcal populations, particularly those from infected tissues. Moreover, this method can be of use in the identification of new Streptococcus species that are potentially involved in polymicrobial infections.
Introduction
Streptococci are anaerobic gram-positive cocci that form a heterogeneous group including more than 30 different species, most of them members of the normal human flora or isolated from other animals. Some species are important human pathogens, like S. pneumoniae, S. pyogenes and S. agalactiae. S. pneumoniae is recognized as a major cause of pneumonia (Citation1); however, S. pneumoniae also causes infections like acute sinusitis, septic arthritis, endocarditis, peritonitis, pericarditis, and severe infections like meningitis or septicaemia. S. pyogenes causes severe infections when it becomes invasive (sepsis, necrotizing fasciitis, etc.), infections that are not severe (tonsillitis, superficial skin infections) and immunological complications (Citation2–4). Finally, S. agalactiae is the most common cause of neonatal sepsis. In addition to these species, S. bovis is occasionally identified as the causative agent in cases of human endocarditis (Citation5) or neonatal septicaemia and meningitis and is found in infections associated with colorectal cancer (Citation6–9) in approximately 25% of cases.
In the last decade, a number of polymerase chain reaction (PCR) assays have been developed for the detection or identification of particular species of this genus. Methods for the specific detection of S. bovis (Citation10), S. agalactiae (Citation11), S. mutans (Citation12) and S. pneumoniae (Citation13) are found in the literature. A PCR targeting the gene tuf has recently been developed to identify the Streptococcus species from clinical isolates (Citation14). However, techniques capable of differentiating and recognizing both cultivatable and non-cultivatable streptococci have not been available.
Culture-independent molecular techniques used in microbial ecology, such as PCR combined with denaturing gradient gel electrophoresis (DGGE), allow the routine survey of entire bacterial communities (Citation15). Phylogenetic analysis of streptococci using several conserved genes coding for heat-shock proteins, 16S rRNA gene, glucose pyrophosphorylase or superoxide dismutase have already been reported, and all of these analyses have demonstrated the usefulness of genetic approaches for improving strepto-coccal identification. We have developed a new 16S rRNA gene primer set specific for the Streptococcus genus and suitable for use in the PCR-DGGE methodological approach. The PCR-DGGE method has been used previously to obtain visual fingerprints of Lactobacillus spp. present in the human intestine (Citation16) or the oral microbial community (Citation17). The primary aim of the present study was the development of a PCR-DGGE-based method for investigating the diversity of the genus Streptococcus in clinical samples.
Material and methods
Microorganisms
A total of 46 bacterial strains were used in this study (), including 17 Streptococcus species covering all major taxonomic groups to date plus 29 other bacterial species covering a wide phylo-genetic range. The strains were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany), from the Spanish Cell Type Collection (Valencia, Spain) and from the microbiology laboratory of the University of Girona.
Table I. Bacterial strains used in the study.
DNA isolation
Bacterial DNA samples were isolated and purified using the commercial DNA extraction kit, DNA genomic Wizard® (Promega, Madison, WI, USA), according to the manufacturer's guidelines for gram-positive bacteria. DNA was stored at –20°C until use.
Clinical samples
The new primer set was tested on six colon biopsies (three healthy controls, samples 10, 13 and 19; and three colorectal cancer patients, samples 4, 16 and 21) and on three bronchoalveolar lavages (samples 55, 59 and 60) from patients admitted to the Intensive Care Unit with severe community-acquired pneumonia. Patients did not receive antibiotic therapy during the 3 months before sample extraction. Samples were provided from the University Hospital Dr Josep Trueta (Girona, Spain) and from the Hospital Santa Caterina (Salt, Spain).
DNA isolation from clinical samples
Biopsy samples were subjected to two mild ultrasound-wash cycles of 45 s each. One of the cycles was performed to discard both transient and loosely attached bacteria. During this process, biopsies were washed with 1 ml of phosphate-buffered saline at pH 7.4. Afterwards, tissue samples were resus-pended in 20 mM Tris/HCl, 2 nM EDTA, 1% Triton X-100, pH 8, with 20 mg/ml lysozyme for 45 min at 37°C. After this step DNA from both biopsies and bronchoalveolar lavages was isolated using the DNA purification kit, Nucleospin® Tissue (Mach-erey & Nagel, Duren, Germany), following the manufacturer's indications for gram-positive bacterial DNA. Purified genomic DNA was stored at –20°C until use.
Primer design
The 16S rRNA gene sequences from all Streptococcus species available in GenBank were aligned using Clustal X 1.81 (Citation18). The following sequences were used: S. acidominimus (accession no. X58301), S. macacae (X58302), S. mutans (X58303), S. rattus (X58304), S. cricetus (X58305), S. downe (X58306), S. sobrinus (X58307), S. oralis (X58308), S. constel-latus (X58310), S. intermedius (X58311), S. pneumo-niae (X58312), S. hyointestinalis (X58313), S. equi (X58314), S. porcinus (X58315), S. iniae (X58316), S. bovis (X58317), S. equinus (X58318), S. alactolyt-icus (X58319), S. salivarius (X58320), S. vestibularis (X58321), S. pyogenes (AM295007), S. sanguinis SK36 (NC009009), S. thermophilus CNRZ1066 (NC006449), S. parauberis strain SAP 9 (AF284579), S. vestibularis strain ATCC 49124 (AY188353), S. macacae strain ATCC 35911 (AY188351), S. sobrinus strain ATCC 33478 (AY188349), S. cristatus strain ATCC 51100 (AY584476), S. waiu (AF088900), S. parasanguis (AF003933), S. anginosus (X58309) and S. mitis (AF003929). Consensus sequences differing from other phylogenetic groups were used to design candidate primers. Resulting oligonucleotides were then tested for specificity using BLAST (Citation19) short sequences mode.
PCR primer suitability was analysed using NetPrimer® and Primer Express (version 2.0) software. Primers were synthesized by TIB MOLBIOL (Berlin, Germany). Primers used in this work are shown in .
Table II. Primers used in this study.
PCR conditions
PCR was performed in a thermal cycler (GeneAmp PCR system 2700, Applied Biosystems, Foster City, CA, USA) adding 1 μl of genomic DNA as a template to a 24 μl PCR mixture containing 1×buffer (II) (Applied Biosystems), 2 mM MgCl2, 200 μM deoxyribonucleoside triphosphates (Universal Master Mix, Applied Biosystems-Roche Molecular Systems Inc., Branchburg, Germany), 500 nM of each primer and 1 U Ampli7agGold™ (Applied Biosystems). The optimal annealing temperature was assayed for the different sets of primers using a gradient thermal cycler (Biometra T Gradient, Göttingen, Germany). For the chosen oligonucleotides (1043F and 1492R) the reaction was conducted as follows: 10 min at 95°C followed by 30 cycles of 30 s at 94°C, 1 min at 67°C and 1 min at 72°C, with a final extension of 10 min at 72°C. Amplification products were analysed by gel electrophoresis through 1.5% agarose gels in 0.5× TBE buffer (45 mM Tris-borate, pH 8.0, 1 mM EDTA) followed by staining with ethidium bromide (0.5 μg/ml).
The amplification limit of the primer set was assayed over a fivefold serial dilution of genomic DNA from S. pneumoniae. Non-template controls were performed to verify the absence of contamination. Furthermore, strict precautions to prevent carry-over of amplified DNA were used after the PCR was completed. The specificity of the primer set was analysed using DNA from the bacteria listed in . Differences in the detection between bacterial samples were evaluated using the Probit test (Minitab® 14 Statistical Software, Pennsylvania, USA). All p values < 0.05 were two-tailed.
DGGE
A 50 GC clamp was added to the 5’ end of the primer 1043F to further separate fragments with denaturing gradient gel electrophoresis (Citation15). DGGE was performed on an Ingeny phorU DGGE system (Vlissingen, the Netherlands). The 6% polyacrylamide gel contained a vertical denaturing gradient ranging from 40% to 70% urea/formamide. Approximately 2 μg DNA was loaded in each lane. Electrophoresis was performed at 60°C in 1× TAE buffer at 120 V for 16 h. The gel was then stained with SYBR gold (Molecular Probes, Paisley, UK) for 30 min, placed on a UV transilluminator and photographed. To standardize the relative positions of bands, two lanes of each gel contained a mix of five 16S rRNA gene fragments from different known species.
Sequencing and analysis of individual DGGE bands
Individual bands were excised with a sterile scalpel and incubated for 45 min in sterile, DNA-free tubes containing 50 μl of 10 mM Tris-HCl, pH 8.0, at 65°C with vigorous vortexing every 5 min to mediate passive diffusion. Supernatants were used as template DNA to generate PCR products of the desired band using the PCR conditions described above with the forward primer 1043F without the GC clamp. PCR products were cleaned and sequenced in both directions (forward and reverse) by Macrogen Inc. (Seoul, Korea) with an ABI 3730CL Automatic DNA sequencer. Sequence alignment was carried out with ClustalW software (Citation18). Consensus sequences were obtained and manually refined with the Bioedit software package (Citation20). Sequences were compared against the National Center for Biotechnology Information (NCBI) database using the BLAST® software (Citation19) and against the Sequence Match software implemented in the Ribosomal Database Project II website (Citation21). Chimeric sequences were checked using the CHECK_CHIMERA program.
GenBank accession numbers
Sequences obtained in this study were deposited in the GenBank database under the accession numbers EU563971 to EU564004.
Results
Streptococcus-specific PCR
A multiple alignment of the 16S rRNA gene sequences from Streptococcus species revealed regions conserved among the streptococci but distinct from those of other bacteria. These conserved 16S rRNA gene sequence regions were used for primer design.
Two primers (623F/R and 1043F/R) were found to be specific for the Streptococcus genus and were combined with the universal eubacterial primers 27F (Citation22), 357F (Citation15) and 1492R (Citation23). Four primer sets were designed in the study. Set 1 was formed by oligonucleotides 27F and 623R, set 2 by 357F and 1043R, set 3 by 623F and 1043R and set 4 by 1043F and 1492R.
After testing the four primer sets designed, it was found that the amplicons generated by set 1 exceeded the expected size and set 3 did not work. The other two primer sets produced amplicons of the expected size. However, since DGGE does not work properly for PCR products larger than 600 bp and set 2 produced a fragment approximately 690 bp in size, set 4 (450 bp) was chosen. Moreover, set 4 enclosed the hypervariable regions V7-V9 that in principle provide a higher resolution for the identification of the fragments. The optimal annealing temperature for this primer pair was empirically determined to be 67°C.
The combination of primers 1043F and 1492R was highly specific for identification of the Streptococcus genus. The 30-cycle PCR assays clearly amplified genomic DNA from the 17 streptococcal species tested. Enterococcus faecalis was the only non-streptococcal bacterial species amplified by the assay (). Specificity was later confirmed with 27 additional Streptococcus isolates from our laboratory.
Table III. Specificity test assay for primers 1043F-1492R on bacterial species from the genusStreptococcus and other phylogenetic groups.
The lowest detectable concentration of template was 170 genome equivalents per reaction (95% probability) after analysing five replicates of a serially diluted S. pneumoniae DNA sample.
Use of primers 1043F-1492R on clinical samples
To test our primer set on clinical samples, we used DNA isolated from mucosal colon biopsies of healthy controls, from patients with colorectal cancer and from bronchoalveolar lavages of patients with streptococcal infection.
The banding pattern obtained by DGGE () represented the principal streptococcal constituents of the analysed community that contribute to > 1% of the total streptococcal population (Citation15). Bands with unclear sequences as the result of having more than one sequence were not obtained.
Figure 1. DGGE profiles of 16S rRNA gene fragments from Streptococcus spp. from colon biopsies (A) and bronchoalveolar lavage samples (B). White dots indicate bands further subjected to re-amplification and sequencing. The percentage of urea + formamide solution is indicated (far right)
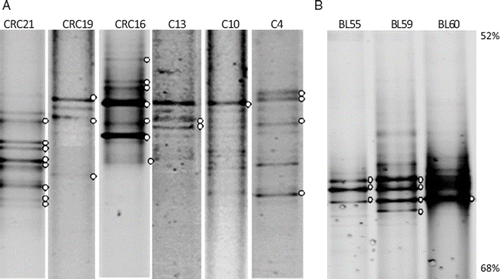
Thirty-three selected 16S rRNA gene PCR products from DGGE bands were sequenced and compared with entries in the Genbank database and Ribosomal Database Project II (). Chimeric sequences were identified by CHECK-CHIMERA software. In the colonic mucosal samples, the dominant phylotypes belonged to the viridans group of streptococci. Sequences matched those of known Streptococcus species such as: S. thermophilus, S. mitis, S. intermedius or S. pneumoniae. Bronchoalveolar lavage samples contained mainly S. pneumoniae. Other streptococci species such as S. thermophilus or S. iniae were also found as accompanying species, which unlike colonic samples belong to the salivarus and pyogenic group. A total of 11 bands had no identity to known cultured strains and probably are non-culturable streptococci. Of these, bands 4a, 13b and 16c corresponded to novel phylotypes as they shared < 94% identity with any GenBank sequences.
Table IV. Sequence identification of DGGE bands in GenBank and RDP II.
Discussion
Streptococci can be found in human, animal, environmental and food sources. Conventional identification methods take 24–48 h after isolation and are often non-conclusive, since some phenotypic criteria cannot always discern between strains or between members of other genera like Enterococcus and Lactococcus. Moreover, many culture procedures overlook a number of viable but non-cultivatable (VBNC) species. In this quiescent state, bacteria are still viable and show metabolic activity, but cannot be shown as colony-forming units by the conventional plate counts. It is estimated that < 2% of bacteria can be cultured in the laboratory, although this is highly variable depending on the environment. For example, about 50% of the oral microbiota have not yet been cultivated and are responsible for several oral infections, and for other body sites, the figure is unknown but is likely to be similar to that found in the mouth or higher (Citation24). Species like Treponema pallidum remain unculturable today and many pathogenic bacteria like Vibrio cholerae, Mycobacterium tuberculosis, Campylobacter jejuni, Helicobacter pylori, Vibrio vulnificus and Escherichia coli have been reported to enter a VBNC state from which they are able to return to the infectious state after passaging in animal hosts (Citation25). Unculturable streptococci can mislead diagnosis when a streptococcal infection is produced.
In recent years molecular methods have provided a fast way to study and identify cultured streptococci (Citation12,Citation26,Citation27). For example, clinically relevant streptococ-cal species can be detected by PCR assays using primers targeting the tuf gene (Citation14). However, tools for the analysis of uncultured streptococcal species have not been developed to date. We have designed a new primer set targeting the small ribosomal subunit gene, which flanks three hypervariable regions (V7, V8 and V9). This primer set can be used in combination with the DGGE technique to retrieve sequences of most streptococci, either cultured or not, from com posite samples.
The results of this study demonstrate that the new primer set is highly genus-specific in PCR assays and can be used to detect and identify streptococcal species from isolates and clinical samples. The method is highly sensitive and detects up to 170 genomic equivalents (95% probability) per reaction. We found that the new primer set also results in the amplification of Ent. faecalis. The reason for this apparent cross-reactivity lies in the extremely high genetic identity between Ent. faecalis and the genus Streptococcus. In fact, Ent. faecalis has been considered a Streptococcus for decades (Citation28,Citation29), they share up to 93% identity in their respective 16S rRNA sequences, and they are closely related phylogenetically. Picard and co-workers (Citation14) also failed to enclose the genus Streptococcus with a unique genus-specific oligonucleotide pair. In that work, the inclusivity range was extended to include Enterococcus durans and Lactococcus lactis. Since the secondary goal of this study was to investigate all the strep-tococcal species present in a given sample, in our opinion the possibility of sporadically obtaining sequences from Enterococcus does not hamper the outcome of this study. Up to 9% of the sequences obtained after DGGE were more similar to Enterococcus spp.
The preliminary application of the new primer set in combination with DGGE for clinical samples revealed the presence of several Streptococcus species or phylotypes. Up to 91% of the retrieved sequences corresponded to the genus Streptococcus and 9% to other genera (Enterococcus and Clostridium). One-third of the Streptococcus sequences belonged to previously unknown, uncultured phylotypes.
Our preliminary results indicate that cultivatabil-ity of the genus Streptococcus is very high (about two-thirds) compared with other microorganisms. Our method allows the identification of the remaining one-third of the genus, which can be useful to complete the picture in those cases where polymicrobial infection is suspected.
The results are consistent with the type of sample analysed. Thus, as expected, S. pneumoniae was found in the three bronchoalveolar specimens studied. In addition, other phylotypes were found to be at least as abundant as S. pneumoniae. The different species in the mitis group can be separated because of the differences in the G+C content of the 16S rRNA gene fragment, whose behaviour in the denaturing gel allows separation and further identification. Our PCR method has a high threshold of approximately 170 genomic equivalents per reaction. Since we add 1 μl from a 50 μl extract of a 1 ml sample volume, the abundance threshold to be detected by PCR is about 8.5 × 103 cells ml-1 if we consider a DNA extraction efficiency of 100%. With this PCR method, the presence of other Streptococcus spp. potentially involved in the ongoing infection can be detected. Although the role of these phylotypes is still unclear, they might be either saprophytic organisms opportunistically growing over the pathogenic activity of S. pneumoniae or additional pathogenic organisms, which may bring other species of clinical interest to light.
As far as the colonic biopsies are concerned, we have identified an unexpected streptococcal diversity, particularly when compared with the diversity of this genus obtained by other authors using generic eubac-terial 16S rRNA gene targeting primers (Citation30–32). This suggests that the study of particular taxonomic or phylogenetic groups requires the design and use of specific primer sets for a better approach to their actual diversity.
Acknowledgements
We thank Dr Xavier Aldeguer and Dr José Maria Sirvent of the Endoscopy Service and ICU of Hospital Josep Trueta, respectively, and Dr Carles Lopez from the Hospital Santa Caterina for kindly providing samples. Drs Marga Martinez and Laia Calvó are acknowledged for critically reading the manuscript.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Whitney CN, Farley MM, Hadler J, Harrison LH, Lexau C, Reingold A, . Increasing prevalence of multidrug resistant Streptococcus pneumoniae in the United States. N Engl J Med 1999; 343:1917–24.
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol 2000; 13:470–511.
- Facklam R, Sahm DS, Teixeira LM. 1999. Streptococci. Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken, RH, Manual of clinical microbiology. Washington, DC: ASM Press. Chapter 17.
- Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, . Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 2007;189:38–51.
- Ruoff KL, Miller SI, Garner CV, Ferraro MJ, Calderwood SB. Bacteremia with Streptococcus bovis and Streptococcus salivarius: clinical correlates of more accurate identification of isolates. J Clin Microbiol 1989;27:305–8.
- Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, . Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis 2000;21:753–6.
- Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Association of Streptococcus bovis with carcinoma of the colon. N Engl J Med 1977;297:800–2.
- Biarc J, Nguyen IS, Pini A, Gosse F, Richert S, Thierse D, . Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S. bovis). Carcinogenesis 2004;25:1477–84.
- Gold JS, Bayar S, Salem RR. Association of Streptococcus bovis bacteremia with colonic neoplasia and extracolonic malignancy. Arch Surg 2004;139:760–5.
- Lee RA, Woo PC, To AP, Lau SK, Wong SS, Yuen KY. Geographical difference of disease association in Streptococcus bovis bacteremia. J Med Microbiol 2003;52:903–8.
- Betriu C, Culebras E, Gomez M, Rodríguez-Avial I, Sanchez BA, Agreda MC, . Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrob Agents Chemother 2003;47:1112–4.
- Chen Z, Saxena D, Caufield PW, Ge Y, Wang M, Li Y. Development of species-specific primers for detection of Streptococcus mutans in mixed bacterial samples. FEMS Microbiol Lett 2007;272:154–62.
- Kresken M, Henrichfreise B, Bagel S, Brauers J, Wiedemann B. High prevalence of the ermB Gene among erythromycin-resistant Streptococcus pneumoniae isolates in Germany during the winter. Antimicrob Agents Chemother 2004;48:3193–5.
- Picard FJ, Ke D, Boudreau DK, Boissinot M, Huletsky A, Richard D, . Use of tuf sequences for genus-specific PCR detection and phylogenetic analysis of 28 streptococcal species. J Clin Microbiol 2004;42:3686–95.
- Muyzer G, deWaal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel elec-trophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 1993;59:695–700.
- Heilig GHJ, Zoetendal EG, Vaughan E, Marteau P, Akkermans ADL, de Vos WM. Molecular diversity of Lacto-bacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 2002;68:114–23.
- Li Y, Ku CYS, Xu J, Sacena D, Caufield PW. Survey of oral microbiota diversity using PCR-based denaturing gradient gel electrophoresis. J Dent Res 2005;84:559–64.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876–82.
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, . Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–402.
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999;41:95–8.
- Larsen N, Olsen GJ, Maidak B, McCaughey L, Overbeek R, Macke TJ, . The ribosomal database project. Nucleic Acids Res 1993;21:3021–3.
- Wang X, Heazlewood SP, Krause DO, Florin TH. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol 2003;95508–20.
- Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol 2002;46:535–48.
- Wade W. Unculturable bacteria – the uncharacterized organisms that cause oral infections. J R Soc Med 2002;95:81–3.
- Yogita N. Viable but non-culturable bacteria: their impact on public health. Curr Science 2005;89:1650
- Kawamura Y, Whiley R, Shu S, Ezaki T, Hardie J. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology 1999;145 (Pt 9): 2605–13.
- Kawamura Y, Hou X, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 1995;45:406–8 published erratum appears in Int J Syst Bacteriol 1995; 45:882.
- Köhler W. The present state of species within the genera Streptococcus and Enterococcus. Int J Med Microbiol 2007;297:133–50.
- Schleifer R, Kilpper-Balz K. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int J Syst Bacteriol 1984;34:31–4.
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, . Diversity of the human intestinal micro-bial flora. Science 2005;308:1635–8.
- Green GL, Brostoff J, Hudspith B, Michael M, Mylonaki M, Rayment N, . Molecular characterization of the bacteria adherent to human colorectal mucosa. J Appl Microbiol 2006;100:460–9.
- Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis 2006;12:1136–45.
