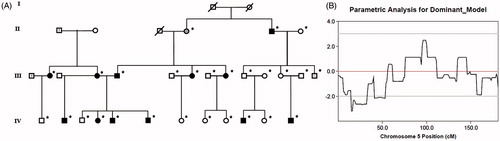
Previously we reported on a two-stage genome-wide linkage scan using microsatellite markers in a four-generation Caucasian family with autosomal-dominant keratoconusCitation1 which identified a peak of 8.2 MB with Lod score of 3.53 located at the 5q14.1-q21.1 chromosomal region.Citation2 To confirm this linkage region we performed a second scan using high density single nucleotide polymorphisms (SNPs) and tested them for genetic linkage and association with keratoconus in 27 family members including those diagnosed with keratoconus denoted by black symbols (). The study was approved by the Institutional Review Board (IRB) and conducted in accordance with the provisions of the Declaration of Helsinki. A total of 525K SNPs (HumanOmniExpress BeadChip, Illumina, Inc.) were tested for linkage under an autosomal dominant model with the same parameters (phenocopy rate of 0.01 and penetrance of 0.5) and pedigree-splitting method as was described in our original paperCitation2 using Merlin 1.1.2.Citation3 A maximum Lod score of 2.49 was calculated at approximately 99 MB region of chromosome 5 overlapping the original linkage peak (). In addition, based on Lod scores greater than 2 we narrowed down the linkage region to 5MB between 95 MB and 100 MB located at 5q15-5q21.1.
FIGURE 1. (A) Family structure of the four generation Caucasian family with keratoconus. *Denotes genotyped individuals. (B) Results of parametric linkage analysis.
We also tested for genetic association 445 SNPs located in this region using generalized estimation equation models accounting for familial correlations (GWAF).Citation4 Association tests under the dominant model identified 32 SNPs with p values ranging from 0.05–0.004; three associated SNPs were located within 50 KB of known genes. These SNPs were rs3853203 (p = 0.01), rs39597 (p = 0.03), and rs11738579 (p = 0.05) located in or adjacent to genes CAST, RIOK2, and RHOBTB3, respectively. All three genes were present in the human keratoconus cornea cDNA library constructed by our group;Citation5 however, variants in CAST gene were implicated in genetic susceptibility to keratoconus in family and case-control panels by our groupCitation6 making it a top candidate gene in the region.
In short, confirmation of this linkage locus makes 5q chromosomal region the strongest candidate for potential identification of a genetic defect associated with familiar nonsyndromic keratoconus in this family as well as possibly in other families with familiar keratoconus.Citation7,Citation8
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
This work was supported by National Eye Institute grant NEI 09052 and the Eye Defects Research Foundation Inc. (to Y.S.R.); NCRR grant M01-RR00425 and Southern California Diabetes Endocrinology Research Center grant DK063491 (to K.D.T).
References
- Rabinowitz YS, Maumenee IH, Lundergan MK, et al. Molecular genetic analysis in autosomal dominant keratoconus. Cornea 1992;11:302–308
- Tang YG, Rabinowitz YS, Taylor KD, et al. Genomewide linkage scan in a multigeneration Caucasian pedigree identifies a novel locus for keratoconus on chromosome 5q14.3-q21.1. Genet Med 2005;7:397–405
- Abecasis GR, Cherny SS, Cookson WO, et al. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002;30:97–101
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics 2010;26:580–581
- Rabinowitz YS, Dong L, Wistow G. Gene expression profile studies of human keratoconus cornea for NEIBank: a novel cornea-expressed gene and the absence of transcripts for aquaporin 5. Invest Ophthalmol Vis Sci 2005;46:1239–1246
- Li X, Bykhovskaya Y, Tang YG, et al. An association between the calpastatin (CAST) gene and keratoconus. Cornea 2013;32:696–701
- Li X, Rabinowitz YS, Tang YG, et al. Two-stage genome-wide linkage scan in keratoconus sib pair families. Invest Ophthalmol Vis Sci 2006;47:3791–3795
- Bisceglia L, De Bonis P, Pizzicoli C, et al. Linkage analysis in keratoconus: replication of locus 5q21.2 and identification of other suggestive loci. Invest Ophthalmol Vis Sci 2009;50:1081–1086

