Axenfeld-Rieger syndrome (ARS) is a rare autosomal dominant disease mainly characterized by maldevelopment of the anterior segment of the eyes, accompanied with dental anomalies and redundant periumbilical skin.Citation1 Some patients have cranial abnormalities with hypoplasia of the midface and an underdeveloped premaxilla, while a few others have hypospadias and growth retardation. A variety of other abnormalities such as heart, hearing and limb defects have also been reported in ARS patients.Citation2 Several transcription factor-encoding genes underlying classic ARS have been identified, among which the pituitary homeobox 2 gene (PITX2) and the forkhead box C1 gene (FOXC1) are most studied.Citation3 FOXC1 is located at 6p25 and is a member of a large superfamily of Forkhead transcription factors, which can recognize and bind to specific DNA sequences through its forkhead domain (FHD). Extraocular anomalies are considered more often in intragenic PITX2 than in intragenic FOXC1 mutations, and larger numbers of individuals with PITX2 mutations have systemic malformation affecting parts of the body other than the eye.Citation2,Citation4 Rare cases of ARS with heart defects caused by the mutation of FOXC1 were reported.
A family from Central-South China with three affected members across three generations participated in the present study. Three patients were diagnosed with ARS and congestive heart abnormalities (). The proband of the family was diagnosed by transthoracic echocardiograms of having congestive heart failure and by gonioscopy of having ARS. No other malformations were observed in the three affected members, indicating this family is an oligosymptomatic ARS family with autosomal dominant pattern. Therefore we examined the possibility of known causative genes that cause ARS. By sequencing analysis of FOXC1 and PITX2, a novel non-synonymous sequence variant, c.380T > G (p.R127L) in the exon of FOXC1 was detected and co-segregated with the ARS phenotye. This newly identified mutation was not found in our 200 control cohortsCitation5 and not presented in the dbSNP and Exome Variant Server database (http://evs.gs.washington.edu/EVS/). Alignment of FOXC1 amino acid sequences from human, mouse, rat, zebrafish etc revealed that the affected amino acid was evolutionarily conserved. Three programs for analyzing protein functions – Polyphen2, SIFT and MutationTaster – predicted that the variants p.R127L are probably damaging, deleterious and disease causing respectively ().
FIGURE 1. Clinical features of the family affected with ARS. (A) Symbols for affected individuals are colored in. The proband (III4) has congenital glaucoma and accompanying congenital heart disease due to PDA. His father (II3) only has glaucoma. His sister (III3) who died in 2010 due to dilated cardiomyopathy, heart failure, cardiogenic shock and accompanying pulmonary edema. (B) Sequence chromatogram indicates a G to T transition of nucleotide 127 in the proband (III4) and his father (II3). (C) The proband’s father has a congenital detachment of the retina as well as glaucoma. (D) The proband also suffers from glaucoma but neither of them have other facial anomalies.
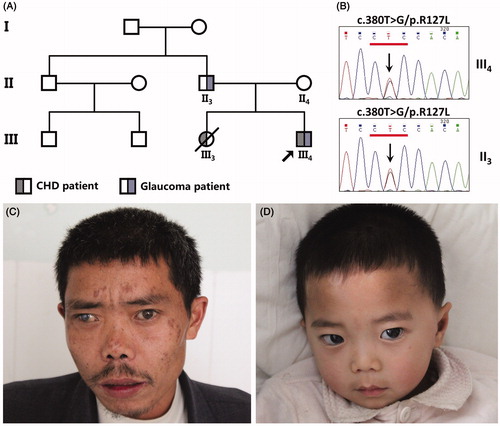
TABLE 1. Summary of identified FOXC1 defects with cardiac anomalies in Axenfeld–Rieger syndrome.
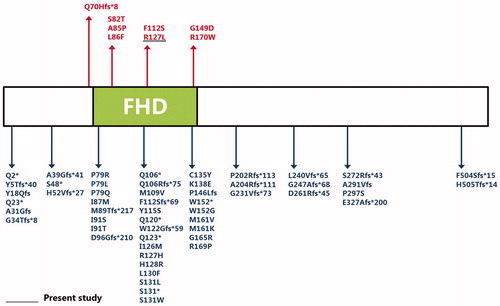
The forkhead box (FOX) proteins are multifaceted transcription factors that are responsible for fine-tuning the spatial and temporal expression of a wide range of genes both in adult tissues and during development, especially in arterial cell specification, angiogenesis and cardiac outflow tract development. All forkhead transcription factors share a highly conserved forkhead DNA-binding domain (FHD) which binds to conserved sequences in the target genes. This domain is composed of one minor and three major alpha-helixes, and two beta-sheets, giving the FHD its characteristic winglike structure. Now we find the novel mutated p.R127L is just located at the FHD. The mutation from alkaline amino acid Arginine positive charge to neutral amino acid Leucine will reduce the positive electrostatic potential on protein surface and disrupt the structure and function of FHD (). Since a host of FOX family proteins are shown to be connected with some heart development-related transcription factors, e.g. NKX2.5, GATA4 etc, we reviewed all of the known mutated sites of FOXC1 (, ),Citation6–16 and noted the sites of heart disease-related mutations, finding some possible common features among them. Interestingly, almost all the heart disease-related mutations are located at the FHD, suggesting that FOXC1 may take an important part in heart development-related pathways through FHD, and any functional damage to FHD is likely to disturb normal heart development.
FIGURE 2. Molecular models of the FOXC1 R127L mutation. (A, B) Diagrams displaying the position of Arg127 and Leu127 (both in yellow), respectively. Red dashed lines show hydrogen bonds. (A) Arg127 linked with Tyr105, Gln123 and Asn124 (both in green) by hydrogen bonds. But (B) Leu127 can only link with Asn124 and Leu130 (both in green) by a hydrogen bond. (C, D) Surface electrostatic charge distribution of FOXC1. As the color legend indicates, the red color (negative potential) arises from an excess of negative charges near the surface and the blue color (positive potential) occurs when the surface is positively charged. Obviously, the alteration of Arg to Leu in 127 reduces the positive potential. Molecular diagrams were drawn with PyMOL (http://www.pymol.org).
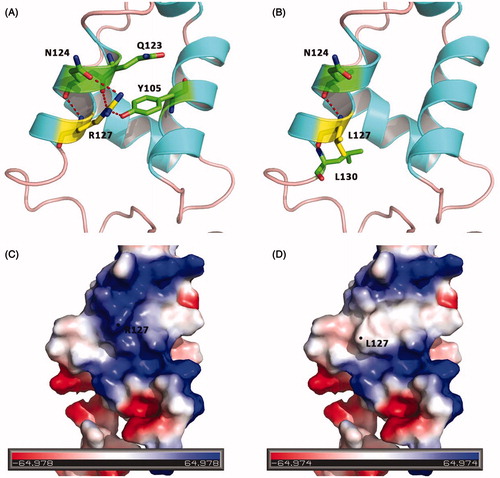
FIGURE 3. Overview of all known and novel FOXC1 mutations, deletions or duplications. The FOXC1 coding region is shown, with all known FOXC1 mutations associated with (red lettering) and not associated with heart disease (blue lettering). Green rectangle: The DNA-binding FHD.
In conclusion, we report a novel FOXC1 mutation (p. R127L) in a three-generation family with three ARS and heart defect patients. The present identification of a novel mutation not only further supports the important role of transcription factor FOXC1 in CHD and ARS, but also expands the spectrum of FOXC1 mutations and will provide insight into genetic diagnosis and counseling of families with ARS and CHD.
Acknowledgements
We thank the patients and their families for participating in this study. We thank the Center of Clinical Gene Diagnosis and Therapy of the State Key Laboratory of Medical Genetics of China for technical assistance.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
This study was supported by the National Natural Science Foundation of China (81370394) and the National Basic Research Program of China (973 Program) (2012CB517900).
References
- Tumer Z, Bach-Holm D. Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur J Hum Genet 2009;17:1527–1539
- Strungaru MH, Dinu I, Walter MA. Genotype-phenotype correlations in Axenfeld-Rieger malformation and glaucoma patients with FOXC1 and PITX2 mutations. Invest Ophthalmol Vis Sci 2007;48:228–237
- Nishimura DY, Swiderski RE, Alward WL, et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 1998;19:140–147
- Chang TC, Summers CG, Schimmenti LA, Grajewski AL. Axenfeld-Rieger syndrome: new perspectives. Br J Ophthalmol 2012;96:318–322
- Tan ZP, Huang C, Xu ZB, et al. Novel ZFPM2/FOG2 variants in patients with double outlet right ventricle. Clin Genet 2012;82:466–471
- Gould DB, Mears AJ, Pearce WG, Walter MA. Autosomal dominant Axenfeld-Rieger anomaly maps to 6p25. Am J Hum Genet 1997;61:765–768
- Mears AJ, Jordan T, Mirzayans F, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet 1998;63:1316–1328
- Saleem RA, Banerjee-Basu S, Berry FB. Analyses of the effects that disease-causing missense mutations have on the structure and function of the winged-helix protein FOXC1. Am J Hum Genet 2001;68:627–641
- Saleem RA, Banerjee-Basu S, Berry FB, et al. Structural and functional analyses of disease-causing missense mutations in the forkhead domain of FOXC1. Hum Mol Genet 2003;12:2993–3005
- Fuse N, Takahashi K, Yokokura S, Nishida K. Novel mutations in the FOXC1 gene in Japanese patients with Axenfeld-Rieger syndrome. Mol Vis 2007;13:1005–1009
- Saleem RA, Murphy TC, Liebmann JM, Walter MA. Identification and analysis of a novel mutation in the FOXC1 forkhead domain. Invest Ophthalmol Vis Sci 2003;44:4608–4612
- Nishimura DY, Swiderski RE, Alward WL, et al. The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 1998;19:140–147
- Swiderski RE, Reiter RS, Nishimura DY, et al. Expression of the Mf1 gene in developing mouse hearts: implication in the development of human congenital heart defects. Dev Dyn 1999;216:16–27
- Honkanen RA, Nishimura DY, Swiderski RE, et al. A family with Axenfeld-Rieger syndrome and Peters Anomaly caused by a point mutation (Phe112Ser) in the FOXC1 gene. Am J Ophthalmol 2003;135:368–375
- Weisschuh N, Dressler P, Schuettauf F, et al. Novel mutations of FOXC1 and PITX2 in patients with Axenfeld-Rieger malformations. Invest Ophthalmol Vis Sci 2006;47:3846–3852
- Gripp KW, Hopkins E, Jenny K, et al. Cardiac anomalies in Axenfeld-Rieger syndrome due to a novel FOXC1 mutation. Am J Med Genet A 2013;161A:114–119

