Abstract
The pyridinium oxime therapy for treatment of organophosphate poisoning is a well established, but not sufficient method. Recent trends also focus on prophylaxis as a way of preventing even the entrance of organophosphates into the nervous system. One of the possible prophylactic methods is increasing the concentration of butyrylcholinesterase in the blood with the simultaneous administration of butyrylcholinesterase reactivators, when the enzyme is continuously reactivated by oxime. This article summarizes and sets forth the structural differences between butyrylcholinesterase and acetylcholinesterase, essential for the future design of butyrylcholinesterase reactivators. Butyrylcholinesterase lacks the reactivator aromatic binding pocket found in acetylcholinesterase, which is itself a part of the acetylcholinesterase peripheral anionic site. This difference finally renders the current acetylcholinesterase reactivators, when used in butyrylcholinesterase, non-functional.
Introduction
Butyrylcholinesterase (BuChE, E.C. 3.1.1.7) and acetylcholinesterase (AChE, E.C. 3.1.1.8) are the main representatives of the cholinesterase family. The cholinesterases are defined by the facts that they hydrolyze choline esters faster than any other substrates and are inhibited by the natural carbamate alkaloid, physostigmine. A particular organism may contain several cholinesterases, distinct both genetically and catalyticallyCitation1. In humans, BuChE is encoded by only one gene, and can be found in the liver, heart, brain, and blood serum, which is why BuChE also used to be called a serum cholinesterase, or simply cholinesteraseCitation2. BuChE is capable of hydrolyzing a vast number of choline esters, with butyrylcholine being one of the most favorable substrates, although its catalytic properties vary from tissue to tissue and species to speciesCitation3. The function of BuChE in an organism is not yet known, and deficient or missing BuChE does not induce any apparent physiological consequencesCitation1. AChE, encoded by a gene distinct from that of the BuChE gene, is mainly located in the nervous system, red blood cells, and muscles. The apparent and long-known function of AChE is to terminate signal transmission in cholinergic synapses by cleaving the neurotransmitter acetylcholine. There are also speculations about other possible functions of AchECitation1. Both BuChE and AChE appear in various molecular forms as soluble tetramers, dimers, and monomers composed of identical subunits, or as amphiphilic membrane-bound forms, also tetrameric and dimeric, or forms immobilized on the collagen tail composed of up to dodekameric assembliesCitation4.
Another characteristic feature of cholinesterases is their inhibition by organophosphate compounds, typically pesticides (parathion, quinalphos, monocrotophos, prophenophos, etc.)Citation5 and chemical warfare agents (tabun, sarin, cyclosarin, soman) in most practical cases. This inhibition, ending with life-threatening poisoning possibly even causing death, is of interest to many research teams throughout the world. The current state-of-the-art post- exposure treatment includes the use of anticholinergic drugs (e.g. atropine), cholinesterase-reactivating agents (usually pyridinium oximes, see ) and anticonvulsant drugs (e.g. benzodiazepines)Citation6.
Figure 1. AChE reactivators. Pralidoxime (2-PAM) as a representative of reactivators with one pyridinium ring and HI-6 as the most typical bisquaternary oxime.
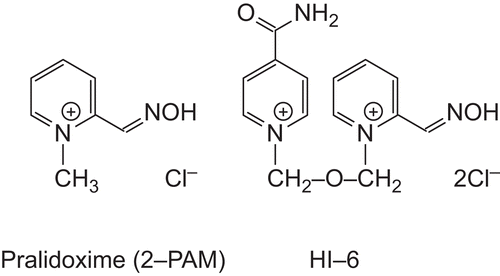
The other approach is to choose an effective prophylaxis. One of the prophylactic therapies suggested is the displacement of organophosphates using a suitable bioscavengerCitation7,Citation8. The bioscavenger of first choice is the human BuChE in combination with oxime reactivators, extending the scavenging capacity beyond the stoichiometric ratio. For this purpose, several AChE reactivators (pralidoxime, HI-6, trimedoxime, etc.) were tested, but their reactivation potency for paraoxon-inhibited BuChE was quite insufficientCitation7. Kinetic studies of the reactivation of tabun-inhibited AChE and BuChE by bisquaternary oximes K024 and K075 revealed their high efficiency in AChE reactivation, but also slow and much less efficient reactivation of BuChECitation9. Pralidoxime is considered a weak reactivator of AchE inhibited by different sarin analogs, and even weaker in the case of BuChECitation10. This effort calls for an exact specification of mutual structural differences between AChE and BuChE for the purposes of rational BuChE reactivator design. The topic of cholinesterase comparison has been addressed several times in past decadesCitation1,Citation4,Citation11–13 using different standpoints: catalysis of natural substrates (choline esters), stereospecificity of enantiomeric alkylphosphonates, reversible inhibition, and substrate inhibition. In the present work, AChE and BuChE have been compared with respect to reactivator binding.
Methods
The primary sequences of BuChE from Homo sapiens and AChE from Mus musculus were extracted from the respective pdb files 1P0ICitation14 and 2GYUCitation15, and the three-letter coded sequence were converted to a one-letter code using the program Three To OneCitation16. A pair-wise alignment was computed by Clustal XCitation17. Alignment parameters were: 10 as the gap opening penalty, 0.1 as the gap extension penalty; and the Gonnet 250 matrix was used as a weight matrix. Furthermore, the three dimensional (3-D) alignment of the crystal structures of BuChE and AChE (1P0I and 2GYU) was done in Swiss-Pdb ViewerCitation18 using the best fit procedure utilizing the backbone atoms. For the purpose of 3-D alignment, all reactivators, carbohydrates, ions, crystallographic agents, and water molecules were deleted from the protein structures.
Results and discussion
As the crystallographic structures of AChE-bound reactivators (see for an example of such a complex) reveal, so far there are three characteristic reactivator binding modes, and the seven most important amino acid residues directly mediating the AChE–reactivator interactions are: Trp286, Tyr124, Tyr72, Glu285, Trp86, Tyr337, and possibly also Gly448 (numbering of residues is taken from the Mus musculus AChE: 2GYU).
Figure 2. Detail of 2GYU active site with the most important residues participating in reactivator binding displayed. The reactivator (HI-6) itself and the catalytic Ser203 are highlighted with bolder tubes.
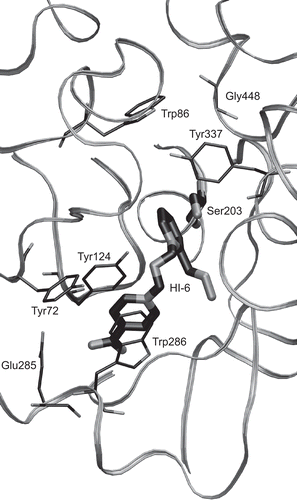
Trp286 stacking with the aromatic ring of a reactivator appears in two of the three reactivator binding modes. In the first mode, the residues stacking with the reactivator ring are Trp286 and Tyr72 from the other side (in 2GYV15, 2GYW15, 2JEZ19, 2JF019); in the second mode the stacking partners are Trp286 and Tyr124 (in 2GYU15, 2JEY19). These two modes are utilized for reactivators with two aromatic rings. The second reactivator ring is located near the catalytic side, and if the aliphatic linking chain connecting two aromatic cores is longer, the second ring interacts with Trp86 and Tyr337. Trp286, Tyr124, and Tyr72, involved in the outer (peripheral) reactivator ring binding, form an aromatic binding pocket. The Glu285 residue located on the bottom of the aromatic pocket electrostatically attracts the positively charged peripheral reactivator ring. Residues of the aromatic binding pocket are part of the peripheral anionic site identified elsewhereCitation20. The third binding mode is exceptionally used by pralidoxime, a reactivator with only one aromatic ring, and the interacting residues are Trp86, Tyr337, and Gly448 (2VQ6). Contrary to other reactivators, pralidoxime is located wholly inside the active site.
An important question remains: are the reactivator binding modes described above for free AchE also valid for binding of reactivators to nerve agent-inhibited AChE? The crystal structures 2JF0 and 2JEZ containing the complexes of tabun-inhibited AChE with Ortho-7 and Hlö-7 suggest that the bisquaternary oximes could utilize the aromatic binding pocket and bind similarly to free AChE and nerve agent-inhibited AChE. The binding need not to be identical, because even Hlö-7 uses different stacking partners for its peripheral ring in the crystallographic structures (Trp286–Tyr124 and Trp286–Tyr72 in the complex with free and inhibited AChE, respectively). The crystal structure of the pralidoxime–AChE complex shows pralidoxime bound on the bottom of the AChE gorge, but nerve agents with bulkier substituents, such as cyclosarin with its cyclohexyl group, will sterically hinder access to the bottom of the gorge. Monoquaternary oximes will probably not bind at the gorge bottom in inhibited AChE complexes.
Identification of reactivator binding modes and residues mediating interactions in AChE allows inspection of the situation for BuChE using different alignment methods. A primary structure alignment () gives overall information about identity (52%) and the similarity of the proteins.
Figure 3. Sequence alignment of BuChE (1P0I) and AChE (2GYU, chain A). The important residues influencing reactivator binding are marked by arrows. The alignment score is represented illustratively by symbols (star, colon, full stop, space) above the alignment rows. The far right column shows the number of the last residue in a row.

Sequences share a high degree of identity and a much higher degree of similarity allowing for, e.g., reliable homology modeling of one structure with use of another structure as a template. This can be also documented by the 3-D alignment of protein backbones (), which are practically overlapping and thus imply that both structures share the same fold.
Figure 4. Aligned protein backbones of BuChE (1P0I) and AChE (2GYU) in rainbow colors and black, respectively.
The active site detail in reveals the main difference between BuChE and AChE with regard to reactivator binding.
Figure 5. 3-D alignment of BuChE (1P0I) and AChE (2GYU). Only the BuChE backbone is displayed and only residues influencing reactivator binding and catalytic Ser203 are shown. The relevant BuChE residues are highlighted by bolder tubes. Labels follow a pattern: BuChE residue/AChE residue.
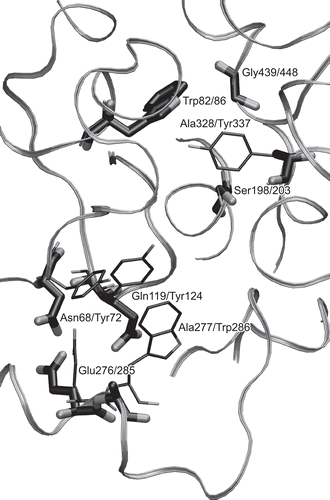
BuChE lacks the aromatic binding pocket found in AChE. The important stacking residues: Trp286, Tyr124, and Tyr72 (2GYU numbering) are replaced by smaller, non-aromatic residues: Ala277, Gln119, and Asn68 (1P0I numbering), respectively, which are not able to stack with aromatic rings, thus preventing the oxime reactivators from efficient binding and performing successful reactivation. The acyl binding pocket of BuChE is much more spacious than the pocket found in AChECitation12, allowing for the cleavage of larger substrates. This fact and the mutation of AChE Tyr337 to Ala238 in BuChE may have further implications for the binding and efficiency of reactivators. Due to the larger active site of BuChE, the dissociation constant of the cyclosarin-inhibited AChE–pralidoxime complex is 50 times higher than the dissociation constant for the cyclosarin-inhibited BuChE–pralidoxime complex, while the values for other sarin analogs are comparableCitation10. In summary, none of the three reactivator binding modes found in crystal AChE structures is directly allowable for reactivator binding in BuChE, making it thus an entirely different case, requiring a distinct treatment and approach in rational reactivator design.
Decleration of interest: This work was supported by the Ministry of Defence of the Czech Republic (KK, DJ), Grant No. FVZ0000604, Ministry of Education of the Czech Republic, Grants No. MSM0021622413 (ZK, JW) and LC06030 (JK).
References
- Massoulié J, Pezzementi L, Bon S, Krejci E, Vallette FM. Molecular and cellular biology of cholinesterases. Prog Neurobiol 1993;41:31–91.
- Lockridge O. Structure of human serum cholinesterase. Bioessays 1988;9:125–8.
- Pauliková I, Hrabovská A, Helia O, Devínsky F. Inter-tissue and inter-species comparison of butyrylcholinesterases. Biologia 2006;61:709–12.
- Chatonnet A, Lockridge O. Comparison of butyrylcholinesterase and acetylcholinesterase. Biochem J 1989;260:625–34.
- Eddleston M, Buckley NA, Eyer P, Dawson AH. Management of acute organophosphorus pesticide poisoning. Lancet 2008;371:597–607.
- Antonijevic B, Stojiljkovic MP. Unequal efficacy of pyridinium oximes in acute organophosphate poisoning. Clin Med Res 2007;5:71–82.
- Jun D, Musilova L, Kuca K, Kassa J, Bajgar J. Potency of several oximes to reactivate human acetylcholinesterase and butyrylcholinesterase inhibited by paraoxon in vitro. Chem Biol Interact 2008;175:421–4.
- Patocka J, Jun D, Bajgar J, Kuca K. Prophylaxis against nerve agent intoxications. Def Sci J 2006;56:775–84.
- Calic M, Bosak A, Kuca K, Kovarik Z. Interactions of butane, but-2-ene or xylene-like linked bispyridinium para-aldoximes with native and tabun-inhibited human cholinesterases. Chem Biol Interact 2008;175:305–8.
- Bartling A, Worek F, Szinicz L, Thiermann H. Enzyme-kinetic investigation of different sarin analogues reacting with human acetylcholinesterase and butyrylcholinesterase. Toxicology 2007;233:166–72.
- Harel M, Sussman JL, Krejci E, Bon S, Chanal P, Massoulié J, et al. Conversion of acetylcholinesterase to butyrylcholinesterase: modeling and mutagenesis. Proc Natl Acad Sci USA 1992;89:10827–31.
- Vellom DC, Radic Z, Li Y, Pickering NA, Camp S, Taylor P. Amino acid residues controlling acetylcholinesterase and butyrylcholinesterase specificity. Biochemistry 1993;32:12–17.
- Taylor P, Radic Z, Hosea NA, Camp S, Marchot P, Berman HA. Structural bases for the specificity of cholinesterase catalysis and inhibition. Toxicol Lett 1995;82–83:453–8.
- Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products. J Biol Chem 2003;278:41141–7.
- Ekstrom F, Pang YP, Boman M, Artursson E, Akfur C, Borjegren S. Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: structural basis for differences in the ability to reactivate tabun conjugates. Biochem Pharm 2006;72:597–607.
- Stothard P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 2000;28:1102–4.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;25:4876–82.
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997;18:2714–23.
- Ekstrom F, Astot C, Pang YP. Novel nerve-agent antidote design based on crystallographic and mass spectrometric analyses of tabun-conjugated acetylcholinesterase in complex with antidotes. Clin Pharmacol Ther 2007;82:282–93.
- Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, et al. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc Natl Acad Sci USA 1993;90:9031–5.
- Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 1993;234:779–815.
