Abstract
A group of 2-alkoxy-5-methoxyallylbenzene were designed, synthesised and evaluated as potential inhibitors of the soybean 15-lipoxygenase (SLO) on the basis of the eugenol and esteragol structures. Compound 4d showed the best half maximal inhibitory concentration (IC50) for SLO inhibition (IC50 = 5.9 ± 0.6 µM). All the compounds were docked in the SLO active site retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB entry: 1IK3) and showed that the allyl group of the synthetic compounds similar to the linoleic acid double bond, were oriented toward the Fe3+-OH moiety in the active site of the enzyme and this conformation was especially fixed by the hydrophobic interaction of the 2-alkoxy group with Leu515, Trp519, Val566 and Ile572. It was concluded that the molecular volume and shape of the alkoxy moiety was a major factor in the inhibitory potency variation of the synthetic compounds.
Keywords::
Introduction
Lipoxygenases (LOs) are non-haem iron-containing enzymes that cause oxidation of polyunsaturated fatty acids and esters to their hydroperoxy derivatives [Citation1]. These families of enzymes widely exist in all plant and animal species [Citation2], and are named according to the oxidised position of the common substrate, arachidonic acid (AA). Among the mammalian lipoxygenases involved in human disease, 5-lipoxygenase (5-LO) is now well documented as a target for reducing the biosynthesis of leukotrienes [Citation3,Citation4]. The 15-lipoxygenase (15-LO) has emerged as an attractive target for therapeutic studies [Citation5], it has been implicated in the progression of certain cancers [Citation6,Citation7] and chronic obstructive pulmonary disease (COPD) [Citation6]. Evidence for the inhibition of 15-LO in the treatment of vascular disease is, however, most compelling [Citation8] and further studies have shown a role for 15-LO in atherogenesis [Citation9,Citation10]. The enzyme is abundantly expressed in macrophages residing within the atherosclerotic lesion [Citation5]. In addition, the immediate products of 15-LO oxidation of AA and linoleic acid (LA) have been shown to be pro-inflammatory [Citation11] and pro-thrombotic [Citation12]. It has also been found that 15-LO is involved in the oxidative modification of low-density lipoproteins (LDL) leading to the development of atherosclerosis [Citation13].
Three different strategies have been developed for inhibiting LO activity [Citation12]. They involve (i) redox inhibitors or antioxidants, which interfere with the redox cycle of 15-LO, (ii) iron-chelator agents, and (iii) non-redox competitive inhibitors, which compete with AA to bind the enzyme active site.
There is reasonable homology between SLO and the human LO [Citation14]. This homology becomes more identical (50%) within 8Å of the active site pocket. The soybean enzyme is much more accessible than the human version and therefore, the results could be extendable to human LO.
Recently we reported the results of our studies on the SLO inhibitory activities of some eugenol esters and on the basis of structure activity relationship (SAR) studies. We suggested that the inhibitory activity of these molecules largely depends on the orientation of the allyl group toward the chelated Fe3+-OH and the molecular volume of the carboxylate moiety in the active site pocket of the enzyme without hydroperoxidation of allylic carbons [Citation14]. In this paper we report the results of a comparative study on the 15-LO inhibitory activities of 2-alkoxy derivatives of 5-methoxyallylbenzene.
Materials and Methods
Chemistry
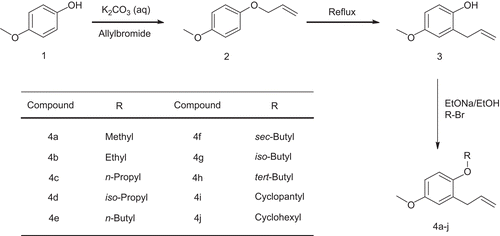
Compound 2 was synthesised by the allylation of 4-methoxyphenol (1) in the presence of saturated potassium carbonate in water [Citation18]. Compound 3 was synthesised using the Claisen rearrangement on compound 2 [Citation19]. The ethers 4b–j were prepared by the reaction of the desired alkylbromides with phenol 3 in the presence of sodium ethoxide in ethanol [Citation20]. Compound 4a and methylisoeugenol were synthesised by the methylation of phenol 3 and isoeugenol [Citation21] using dimethylsulphate [Citation22]. All of the products 4a–j were purified by column chromatography (Silicagel 230–400, eluent: hexane/dichloromethane 50:50).
Molecular modelling, docking and SAR study
Multiple alignment
The conserved amino acids were identified through multiple alignment in clustalX 1.81 [Citation23]. The sequences of the lipoxygenase (LO) family were selected from blasted sequences using the ExPASY proteomics server [Citation24]. The multiple alignment process was then carried out on the selected sequences (protein weight matrix: BLOSUM series, opening gap penalty = 10).
Calculations
The structures of the desired compounds were drawn using the ChemOffice professional (Cambridge software) [Citation25]. The output files were minimised under the molecular mechanic MM+ in HyperChem7.5 [Citation26]. Then the structures were minimised under the semi-empirical AM1 method in the second optimisation (Convergence limit = 1e–5; Iteration limit = 100; RMS gradient = 0.05 kcal/mol; Fletcher-Reeves optimiser algorithm) in HyperChem7.5 [Citation26,Citation27].The crystal structure of the soybean lipoxygenase-3 (arachidonate 15-lipoxygenase) complex with 13(S)-hydroperoxy-9(Z)-2,11(E)-octadecadienoic acid was retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB entry: 1IK3).
Molecular docking
The automated docking simulation was implemented to dock 4a–j into the active site of SLO using the AutoDockTools (ADT) version 1.5.4 [Citation15] with the Lamarckian genetic algorithm [Citation28]. This method has been previously shown to produce bonding models similar to the experimentally observed models [Citation14, Citation28–30]. The torsion angles of the ligands were identified, hydrogens were added to the macromolecule, bond distances were edited and solvent parameters were added to the enzyme 3D structure. Partial atomic charges were then assigned to the macromolecule as well as ligands (Gasteiger for the ligand and Kollman for the protein). The regions of interest of the enzyme were defined by considering the Cartesian chart 20.6, 7.2 and 20.5 as the centre of a grid size of 50, 60 and 60 points in the X, Y and Z axes. The docking parameter files were generated using Genetic Algorithm and Local Search Parameters (GALS) while the number of generations was set to 100. The 100 docked complexes were clustered with a root-mean-square deviation tolerance of 1 Å. Autodock generated 100 docked conformers of 4a–j, corresponding to the lowest-energy structures. After the docking procedures in ADT4, the docking results were submitted to the Weblab Accelrys DS Visualizer 2.0.1 [Citation31] and Swiss-PdbViewer 3.7 (spdbv) [Citation32] for further evaluations. The results of the docking processing (Ki: Estimated Inhibition Constant) are outlined in Table I.
Table I. Enzyme inhibitory assessment and docking analysis data of consensus conformers of eugenol, methyl eugenol, methyl isoeugenol and compounds 4a–j. Ki (M): Estimate d Inhibition Constant. The IC50 (µM) values are given as mean ± SD.
15-LO inhibitory assessment
Lipoxygenase activity was measured in borate buffer solutions (0.1 M, pH 9) using the method described in the literature [Citation33,Citation34], by measuring the absorbance at 234 nm for 60 s after addition of the enzyme (soybean 15-lipoxygenase) and linoleic acid (final concentration: 134 µM) as substrate at 20 ± 1°C. The final enzyme concentration was 167 U/mL. The synthesised substances were added as DMSO solutions (final inhibitor concentrations: 200, 100, 50, 25, 12.5, 6.25, 3.12 and 1.5 µM; final DMSO concentration 1%); whereas DMSO was added in the control experiments with no inhibitor. The mixture of each inhibitor and linoleic acid were set as the blank sample in the testing step. At least six control test tubes and three tubes for each inhibitor solution were measured. To ensure a constant enzyme activity throughout the experiment, the enzyme solution was kept on ice, and the controls were measured at regular intervals. The calculation of enzyme activity was carried out as previously described [Citation34] and the IC50 values were determined by using a sigmoidal dose-response curve in Graphpad Prism 3.02 [Citation35].
Experimental Section
Instruments
Melting points were recorded on an Electrothermal type 9100 melting point apparatus. The 1H NMR (100 MHz) spectra were recorded on a Bruker AC-100 spectrometer. Elemental analysis was obtained on a Thermo Finnigan Flash EA microanalyser. The IR spectra were obtained on a 4300 Shimadzu Spectrometer. All measurements of lipoxygenase activities were carried out using an Agilent 8453 photodiode array spectrophotometer. The soybean 15-lipoxygenase, eugenol, methyleugenol and other chemicals were purchased from Fluka and Merck.
1-Allyloxy-4-methoxybenzene (2)
A mixture of 25.2 g (0.2 mol) of 4-methoxyphenol, 26.4 g (0.22 mol) of allylbromide, and 28 g of potassium carbonate (0.2 mol) in 40 mL water was refluxed for 5 hours and cooled. The mixture was diluted with 100 mL of water and extracted with ether (2 × 50 mL). The combined extracts were washed with 10% NaOH (2 × 50 mL) and dried with anhydrous potassium carbonate. After removal of the solvent, the residual oil was distilled under reduced pressure.
Colourless oil; yield: 78%; bp: 118°C/2 mm: 1HNMR (CDCl3): δ 3.78(s, 3H, -OCH3), 4.49 (d, J = 6.2 Hz, 2H, -CH2-), 5.2–5.52 (m, 2H, H2C=), 5.82–6.25 (m, 1H, HC=), 6.74–6.93 (m, 3H, H-3, H-5, H-6); IR cm−1: 2850 (C=O).
2-Allyl-4-methoxyphenol (3)
The allyl ether 3 (35 g) was refluxed for 3 hours under a nitrogen atmosphere, then cooled. The oil was dissolved in 100 mL of ether and the solution was extracted with 10% NaOH (3 × 50 mL). The combined alkaline extracts were then acidified with 50 mL of concentrated HCl diluted with 50 mL of water, and the mixture was extracted with ether (3 × 50 mL). The combined ether extracts were dried with anhydrous sodium sulphate, evaporated and the residual oil was distilled under reduced pressure.
Colourless oil; yield: 70%; bp: 120°C/2 mm: 1HNMR (CDCl3): δ 3.38 (d, J = 6.2 Hz, 2H, -CH2-), 3.76 (s, 3H, -OCH3), 4.64 (s, 1H, -OH), 5.04–5.25 (m, 2H, H2C=), 5.8–6.26 (m, 1H, HC=), 6.59–6.97 (m, 3H, H-3, H-5, H-6); IR cm−1: 2850 (C=O), 3400 (O-H).
General procedure for preparation of compounds (4a–j)
A mixture of 0.6 g (8.8 mmol) of sodium ethoxide in 5 mL ethanol, 1.4 g (8.8 mmol) of ally ether 3 and 11 mmol of the desired alkyl bromide (for compounds 4a and 4b dimethyl sulphate and ethyl iodide were used, respectively) was refluxed for 1 hour (2 hours for compound 4a). After cooling, the mixture was poured into cold water (10 mL). The mixture was acidified with 20% HCl and then extracted with ether (2 × 10 mL). The combined extracts were washed with 10% NaOH (2 × 10 mL) dried with anhydrous sodium sulphate and evaporated then the residual oil was purified by column chromatography.
2-Allyl-1,4-dimethoxybezene (4a)
Colourless oil; yield: 74%; 1HNMR (CDCl3): δ 3.35 (d, J = 6.2 Hz, 2H, -CH2-), 3.74 (s, 3H, -OCH3), 3.76 (s, 3H, -OCH3), 4.94–5.2 (m, 2H, H2C=), 5.76–6.2 (m, 1H, HC=), 6.62–6.9 (m, 3H, H-3, H-5, H-6); IR cm−1: 2850 (C=O).
2-Allyl-1-ethoxy-4-methoxybenzene (4b)
Colourless oil; yield: 79%; 1HNMR (CDCl3): δ 1.42 (t, J = 6.9 Hz, 3H, -CH3), 3.41 (d, J = 6.2 Hz, 2H, -CH2-), 3.78 (s, 3H, -OCH3), 4.01 (q, J = 6.9 Hz, 2H, -OCH2CH3), 5–5.22 (m, 2H, H2C=), 5.81–6.25 (m, 1H, HC=), 6.62–6.89 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 74.92; H, 8.33. C12H16O2 requires: C, 74.97; H, 8.39%.
2-Allyl-4-methoxy-1-propoxybenzene (4c)
Colourless oil; yield: 75%: 1HNMR (CDCl3): δ 1.05 (t, J = 7.3 Hz, 3H, -CH3), 1.62–2 (m, 2H, -OCH2CH2CH3), 3.39 (d, J = 6.6 Hz, 2H, -CH2-), 3.76 (s, 3H, -OCH3), 3.88 (t, J = 6.3 Hz, 2H, -OCH2CH2CH3), 5–5.23 (m, 2H, H2C=), 5.8–6.23 (m, 1H, HC=), 6.6–6.84 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 75.89; H, 8.83. C13H18O2 requires: C, 75.69; H, 8.80%.
2-Allyl-1-isopropoxy-4-methoxybenzene (4d)
Colourless oil; yield: 50%; 1HNMR (CDCl3): δ 1.32 (d, J = 6.1 Hz, 6H, (H3C)2CHO-), 3.38 (d, J = 6.6 Hz, 2H, -CH2-), 3.75 (s, 3H, -OCH3), 4.25–4.58 (m, 1H, -OCH(CH3)2), 4.97–5.21 (m, 2H, H2C=), 5.78–6.19 (m, 1H, HC=), 6.59–6.87 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 75.77; H, 8.74. C13H18O2 requires: C, 75.69; H, 8.8%.
2-Allyl-1-butoxy-4-methoxybenzene (4e)
Colourless oil; yield: 53%; 1HNMR (CDCl3): δ 0.95 (t, J = 6.8 Hz, 3H, -CH3), 1.23–1.94 (m, 4H, -OCH2(CH2)2CH3), 3.35 (d, J = 6.6 Hz, 2H, -CH2-), 3.74 (s, 3H, -OCH3), 3.89 (t, J = 6.2 Hz, 2H, -OCH2(CH2)2CH3), 4.94–5.18 (m, 2H, H2C=), 5.78–6.21 (m, 1H, HC=), 6.57–6.81 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 76.47; H, 9.14. C14H20O2 requires: C, 76.33; H, 9.15%.
2-Allyl-1-sec-butoxy-4-methoxybenzene (4f)
Colourless oil; yield: 73%; 1HNMR (CDCl3): δ 0.86 (t, J = 7.1 Hz, 3H, -OCH(CH3)CH2CH3), 1.2 (d, J = 6.1 Hz, 3H, -OCH(CH3)CH2CH3), 1.41–1.75 (m, 2H, -OCH(CH3)CH2CH3), 3.74 (s, 3H, -OCH3), 3.89 (m, 1H, -OCH (CH3)CH2CH3), 4.94–5.18 (m, 2H, H2C=), 5.78–6.21 (m, 1H, HC=), 6.57–6.81 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 76.37; H, 9.19. C14H20O2 requires: C, 76.33; H, 9.15%.
2-Allyl-1-isobutoxy-4-methoxybenzene (4g)
Colourless oil; yield: 70%; 1HNMR (CDCl3): δ 1.32 (d, J = 6.1 Hz, 6H, (H3C)2CHO-), 3.38 (d, J = 6.6 Hz, 2H, -CH2-), 3.75 (s, 3H, -OCH3), 4.25–4.58 (m, 1H, -OCH(CH3)2), 4.97–5.21 (m, 2H, H2C=), 5.78–6.19 (m, 1H, HC=), 6.59–6.87 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 76.27; H, 9.1. C14H20O2 requires: C, 76.33; H, 9.15%.
2-Allyl-1- tert-butoxy-4-methoxybenzene (4h)
Colourless oil; yield: 18%; 1HNMR (CDCl3): δ 1.37 (s, 9H, -CH3), 3.39 (d, J = 6.1 Hz, 2H, -CH2-), 3.74 (s, 3H, -OCH3), 4.97–5.27 (m, 2H, H2C=), 5.62–6.21 (m, 1H, HC=), 6.30–7.02 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 76.19; H, 9.09. C14H20O2 requires: C, 76.33; H, 9.15%.
2-Allyl-1-cyclopentyloxy-4-methoxybenzene (4i)
Colourless oil; yield: 65%; 1HNMR (CDCl3): δ 1.57–1.63 (m, 8H, -CH2- (cyclopentyl)), 3.33 (d, J = 6.6 Hz, 2H, -CH2-), 3.75 (s, 3H, -OCH3), 4.82–4.6 (m, 1H, -CH- (cyclopentyl)), 4.97–5.2 (m, 2H, H2C=), 5.77–6.22 (m, 1H, HC=), 6.54–6.74 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 77.45; H, 8.64. C15H20O2 requires: C, 77.55; H, 8.68%.
2-Allyl-1-cyclohexyloxy-4-methoxybenzene (4j)
Colourless oil; yield: 63%: 1HNMR (CDCl3): δ 1.24–2.13 (m, 10H, -CH2- (cyclohexyl)), 3.37 (d, J = 6.6 Hz, 2H, -CH2-), 3.74 (s, 3H, -OCH3), 3.95–4.22 (m, 1H, -CH- (cyclohexyl)), 4.92–5.2 (m, 2H, H2C=), 5.76–6.15 (m, 1H, HC=), 6.54–6.88 (m, 3H, H-3, H-5, H-6); IR cm−1: 2950 (C=O). Found: C, 78.13; H, 9.04. C16H22O2 requires: C, 78.01; H, 9%.
Results and discussion
Considering our previous work on eugenol and esters [Citation14], we tested the inhibitory properties of eugenol, methyl eugenol, methyl isoeugenol and 2,5-dimethoxyallylbenzene on SLO (substrate: linoleic acid) to study the effect of the methoxy group position on the inhibitory activity. The results showed half maximal inhibitory concentration (IC50) of 34.1 ± 2.1, 96.1 ± 3.3, 138.9 ± 3.1 and 93.1 ± 2.9 µM for the above mentioned compounds, respectively (Table I). It was found that the protection of the hydroxyl group of eugenol decreased its IC50 and among the dimethoxy compounds, methyl isoeugenol showed a reduced inhibitory activity. It is notable that no other products such as hydroperoxy were isolated from the action of the LO enzyme on the methylated compounds as substrate (assuming hydroproxy is supposed to be obtained if the redox pathway is blocked and the inhibitor acts through its allylic group in reaction with the enzyme active site similar to the oxidation of natural unsaturated fatty acids).Footnote†
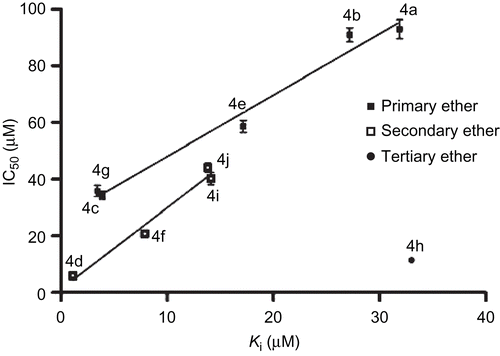
Considering the IC50 results of the above compounds, and approved site direction of the allyl group in the active site pocket of SLO, we decided to synthesis other 2-alkyl analogues of 2,5-dimethoxyallylbenzene to investigate the interactive effect of the 2-alkyl moiety with the lipophilic pocket formed by the conserved residues Leu515, Val566, Leu565 Ile572 and Leu773 on the inhibitory potency. To investigate this, some 2-alkyl and cycloalkyl analogues of 2,5-dimethoxyallylbenzene (compounds 4a–j) were synthesised) and their inhibitory potency was determined. The synthetic ethers 4a–j showed a broad range of inhibition activity on the enzyme (IC50 = 5.9 to 93.1 µM, see Table I). Compound 4d, with an isopropyl substituent, was the most potent inhibitor at 5.9 ± 0.6 µM while the methoxy and ethoxy analogues (4a and 4b) showed less activity (IC50 = 93.1 ± 2.9 and 91.1 ± 3.2 µM, respectively). The secondary ethers (4d, 4f, 4i and 4j) showed the best inhibitory activity in comparison with the primary types. Among the butoxy analogues, the tertiary type (4h) possessed the greatest inhibitory potential with an IC50 value of 11.4 ± 0.6 µM. The results indicated that an increase in the C-O substituent number led to an improvement in inhibitory potency for this series of compounds.
The binding affinity of compounds 4a–j towards SLO was studied. 100 docked conformers of the desired compounds were generated using the AutoDockTools software in which the side chain of Leu515, Val566, Leu565 Ile572 and Leu773 was flexibilised [Citation15]. The experimental results matched with the theoretical Ki for the docking study for those models in which the allylic double bond oriented toward the iron atom in a similar orientation to that found in linoleic acid (LA) in the active site of SLO. One conformer from each ether cluster which had more similarity with the optimum conformer (lowest Ki) of 4d was adopted as the “consensus” structure and used for further analysis [Citation14,Citation16].
Figure 2. Stick (right) and solvent surface (left) view of flexible residues, surrounding the consensus bonding conformations of compounds 4a–j in the SLO active site.
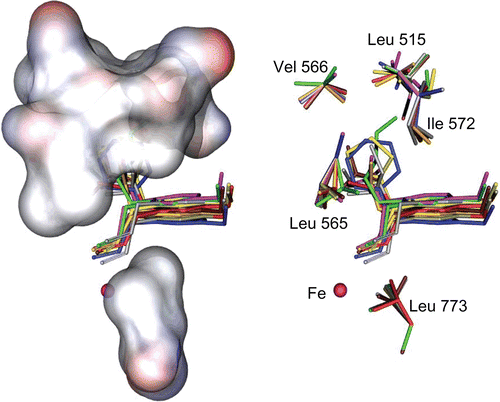
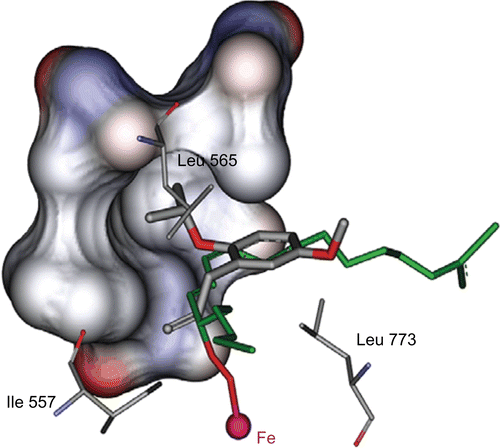
The results of the docking analysis showed that the consensus structure of 4a–j had an estimated inhibitory constant (Ki) in the range of 1 to 33 × 10−6 M (Table I). It appeared that the allyl benzene portion of the consensus structure has a hydrophobic interaction with the backbone of the Trp519, Ile557, Leu565, Ile572, and Leu773 in such an orientation. The Ile557 → Ala and Ile572 → Phe mutants decreased the kcat by two fold from the WT (wild type), While Leu565 → Ala and Leu773 → Ala decreased the kcat by 60 and 1000 fold respectively, indicating that these hydrophobic residues (specially Leu565 and Leu773) contributed significantly to catalysis [Citation17]. Mutating the residues Ile or Leu to Ala, shape an empty space in the binding pocket of SLO, led to a decreased H• transfer kinetics [Citation17]. According to the multiple alignment results, the amino acids Ile557, Leu565 and Leu773 were found to be conserved over all species ().
Figure 3. Multiple alignment of SLO (1ik3_A). The residues which have lipophilic interactions with docked ligands are highlighted in green respectively.

The Ki variations of the consensus structure of the primary and secondary ethers were distinct, in contrast with the IC50 changes (). This relation comes from the tendency of the alkyl moiety to fill the empty lipophilic space formed by the Leu515, Trp519, Val566 and Ile572 side chains. The docking results and IC50 values of the synthetic ethers, indicated that the aforementioned lipophilic cavity of SLO has an organised space for accepting the isopropyl group in comparison with the other structural isomer.
Figure 5. Stick view of the consensus bonding conformation of compound 4d which has lipophilic interaction through its isopropyl moiety with hydrophobic pockets (surface view) formed by Leu515, Trp519, Val566 and Ile572 side chain. In this figure we can see the orientation of linoleic acid (green stick) bonded to Fe via proxy bridge.
In summary, we carried out SAR comparative studies on some methoxyallylbenzene derivatives as 15-lipoxygenase inhibitors. We suggest that the application of docking results such as Ki can be applied to predict the inhibitory potency of the synthetic ethers. This study has also shown the important role of molecular structure in the inhibitory activity of 2-alkoxy-5-methoxyallylbenzene. The importance of these compounds is highlighted for having a straightforward synthesis pathway with a high yield.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Notes
† Substrate (100 μM) was reacted with soybean LO enzyme (167 U/mL) in 3 mL borate buffer solution (0.1 M, pH 9) at 20°C for 15 min. The mixture was then analysed by UV at 230–270 nm and no absorption of vinyl benzene formation was appeared over the blank solution.
References
- Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem 1999;274:23679–23682.
- Kuhn H, Thiele B. The diversity of the lipoxygenase family: Many sequence data but little information on biological significance. J FEBS Lett 1999;449:7–11.
- Larsen JS, Acosta EP. Leukotriene-receptor antagonists and 5-lipoxygenase inhibitors in asthma. Ann Pharmacother 1993;27:898–903.
- Schewe T. 15-lipoxygenase-1: a prooxidant enzyme. Biol Chem 2002;383:365–374.
- Kelavkar U, Glasgow W, Eling TE. The effect of 15-lipoxygenase-1 expression on cancer cells. Curr Urol Rep 2002;3:207–214.
- Kelavkar UP, Cohen C, Kamitani H, Eling TE, Badr KF. Concordant induction of 15-lipoxygenase-1 and mutant p53 expression in human prostate adenocarcinoma: correlation with Gleason staging. Carcinogenesis 2000;21:1777–1787.
- Zhu J, Kilty I, Granger H, Gamble E, Qiu YS, Hattotuwa K, Elston W, Liu WL, Liva A, Pauwels RA, Kis JC, Rose VD, Barnes N, Yeadon M, Jenkinson S, Jeffery PK. Gene expression and immunolocalization of 15-lipoxygenase isozymes in the airway mucosa of smokers with chronic bronchitis. Am J Respir Cell Mol Biol 2002;27:666–677.
- Zhao L, Funk CD. Lipoxygenase pathways in atherogenesis. Trends Cardiovasc Med 2004;14:191–195.
- Cyrus T, Witztum JL, Rader DJ, Tangirala R, Fazio S, Linton, MF, Funk CD. Disruption of the 12/15-lipoxygenase gene diminishes atherosclerosis in apo E-deficient mice. J Clin Invest 1999;1597–1604.
- Harats D, Shaish A, George J, Mulkins M, Kurihara H, Levkovitz H, Sigal E. Overexpression of 15-lipoxygenase in vascular endothelium accelerates early atherosclerosis in LDL receptor-deficient mice. Arteriosler Thromb Vasc Biol 2000;20:2100–2105.
- Sultana C, Shen Y, Rattan V, Kalra VJ. Lipoxygenase metabolites induced expression of adhesion molecules and transendothelial migration of monocyte-like HL-60 cells is linked to protein kinase C activation. J Cell Phys 1996;167:467–487.
- Setty BN, Werner MH, Annun YA, Stuart M. 15-Hydroxyeicosatetraenoic acid-mediated potentiation of thrombin-induced platelet functions occurs via enhanced production of phosphoinositide-derived second messengers--sn-1,2-diacylglycerol and inositol-1,4,5-trisphosphate. J Blood 1992;80:2765–2773.
- Charlier C, Michaux C. Dual inhibition of cyclooxygenase-2 (COX-2) and 5-lipoxygenase (5-LOX) as a new strategy to provide safer non-steroidal anti-inflammatory drugs. Eur J Med Chem 2003;38:645–659.
- Sadeghian H, Seyedi SM, Saberi MR, Arghiani Z, Riazi M. Design and synthesis of eugenol derivatives, as potent 15-lipoxygenase inhibitors. Bioorg Med Chem 2008;16:890–901.
- Auto Dock Tools (ADT), The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037-1000, USA; (http://www.scripps.edu/pub/olson-web/doc/autodock/); Python MFS. A programming language for software integration and development. J Mol Graphics Mod 1999;17:57–61.
- Sadeghian H, Sadeghian A, Pordel M, Rahimizadeh M, Jahandari P, Orafaie A, Bakavoli M. Design, synthesis, and structure–activity relationship study of 5-amido-1-(2,4-dinitrophenyl)-1H-4-pyrazolecarbonitrils as DD-carboxypeptidase/penicillin-binding protein inhibitors with Gram-positive antibacterial activity. Med Chem Res 2010;19:103–119.
- Knapp MJ, Seebeck FP, Klinman JP. Steric Control of Oxygenation Regiochemistry in Soybean Lipoxygenase-1. J Am Chem Soc 2001;123:2931–2932.
- Seyedi SM, Sadeghian H, Rezai M. The green method for regiospecific ring opening of epoxide with dimercaptoethane. Phosphorus Sulfur and Silicon 2007; 182:1709–1716.
- Tarbell DS. The claisen rearrangement. Chem Review 1940;27:495–546.
- Vogel AI. Vogel’s Textbook of Practical Organic Chemistry Langman Scientific & Technical, New York, USA, 1989:413.
- Allen CFH, Gates JW. o-Eugenol. Org Synthesis 1945;25:49–50.
- Vogel AI. Vogel’s Textbook of Practical Organic Chemistry Langman Scientific & Technical, New York, USA, 1989:755.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 1997;24:4876–4882.
- http://us.expasy.org/
- ChemDraw® Ultra, Chemical Structure Drawing Standard, CambridgeSoft Corporation, 100 Cambridge Park Drive, Cambridge, MA 02140 USA, http://www.cambridgesoft.com
- HyperChem® Release 7, Hypercube Inc., http://www.hyper.com/
- Sadeghian H, Attaran N, Jafari Z, Saberi MR, Pordel M, Riazi MM. Design and synthesis of 4-methoxyphenylacetic acid esters as 15-lipoxygenase inhibitors and SAR comparative studies of them. Bioorg Med Chem 2008;17:2327–2335.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical bonding free energy function. J Comput Chem 1998;19:1639–1662.
- Sippl W. Receptor-based 3D-QSAR analysis of estrogen receptor ligands--merging the accuracy of receptor-based alignments with the computational efficiency of ligand-based methods. J Comput Aided Mol Des 2000;14:559–572.
- Dym O, Xenarios I, Ke H, Colicelli J. Molecular docking of competitive phosphodiesterase inhibitors. Mol Pharmacol 2002;61:20–25.
- http://accelrys.com/products/discovery-studio/
- Swiss-pdbViewer 3.6, Glaxo Wellcome Experimental Research, http://www.expasy.org/spdbv/
- Malterud KE, Rydland KM. Inhibitors of 15-lipoxygenase from orange peel. J Agric Food Chem 2000;48:5576–5580.
- Malterud KE, Farbrot TL, Huse AE, Sund R. B. Antioxidant and radical scavenging effects of anthraquinones and anthrones. Pharmacology 1993;47:77–85.
- http://www.graphpad.com/