Abstract
Protein disulphide isomerase (PDI) in the endoplasmic reticulum catalyzes the rearrangement of disulphide bridges during folding of secreted proteins. It binds various molecules that inhibit its activity. But here, we looked for molecules that would potentiate its activity. PDI reductase activity was measured in vitro using di-eosin-oxidized glutathione as substrate. Its classical inhibitor bacitracin was found to exert a biphasic effect: stimulatory at low concentrations (∼10−6 M) and inhibitory only at higher concentrations (∼10−4–10−3 M). The weak oestrogenic molecule bisphenol A was found to exert a weak inhibitory effect on PDI reductase activity relative to the strong oestrogens, ethynylestradiol, and diethylstilbestrol. Like 19-nortestosterone, fluoxetine was found to exert a potentiating effect on PDI reductase activity and their potentiating effects could be reversed by increasing concentrations of oestrogens. In conclusion, this paper provides the first identification of potentiators of PDI activity that are potential pharmaceuticals against pathologies affecting protein folding such as Alzheimer’s disease.
| Abbreviations | ||
| 19-NT, | = | 19-nortestosterone; |
| BAC, | = | bacitracin; |
| BPA, | = | bisphenol A; |
| DES, | = | diethylstilbestrol; |
| DiE-GSSG, | = | di-eosin oxidized glutathione; |
| EE2, | = | ethynylestradiol; |
| FLX, | = | fluoxetine; |
| MPA, | = | medroxyprogesterone acetate |
Introduction
Protein disulphide isomerase (PDI) has been known for a long time as a protein-folding catalystCitation1 involved in the formation of native disulphide bridges of proteins in the endoplasmic reticulumCitation2,Citation3. It binds to numerous ligands including proteins, peptides, and various other molecules. In particular, PDI has been shown to bind oestradiol and this leads to a decrease in its isomerase activity as followed by its activity on RNAse renaturationCitation4.
Using di-eosin oxidized glutathione (DiE-GSSG) as substrate, it has been previously shownCitation5 that all the potent oestrogenic molecules tested [oestradiol, E2; 17α-ethynylestradiol (EE2) and diethylstilbestrol (DES)] also exhibited an inhibitory effect on PDI reductase activity. Likewise, the non-steroidal anti-inflammatory molecule indomethacin was also found to inhibit PDI reductase activity in the same assay.
Surprisingly, we found that two non-oestrogenic steroids, medroxyprogesterone acetate (MPA) and 19-nortestosterone (19-NT), in contrast potentiated PDI reductase activityCitation5. In order to get a better understanding of the ligands properties responsible for inhibition or augmentation of PDI reductase activity, we performed a preliminary screening with various other molecules in order to get a first guess on the molecular structural features of the ligands exhibiting either inhibitory or stimulatory effects, respectively. In addition to the previously identified 19-NT, three other molecules, bacitacin, fluoxetine (FLX) and ammonium sulphate, were found to potentiate PDI reductase activity using DiE-GSSG as substrate.
Materials and methods
Bacitracin (BAC), bisphenol A (BPA), 17α-EE2, FLX, dithioerythreitol (DTeT), eosin 5-isothiocyanate, GSSG, PDI (E.C. 5.3.4.1) from bovine liver (PDI), were all purchased from Sigma–Aldrich (Isle-d’Abeau, France) and were of the highest available grades.
The PDI substrate, DiE-GSSG, was synthesized and purified as previously describedCitation6 with minor modificationsCitation5.
PDI reductase activity was measured through abolishment of fluorescent self quenching when DiE-GSSG is reduced into two molecules of eosin-reduced glutathione (E-GSH). Initial velocities in fluorescence increase (λexc = 518 nm; λem = 545 nm) were recorded using a Spectra-Max Gemini spectrofluorimeter (Molecular Devices, Sunnyvale, California) and analyzed with SoftMaxPro program (Molecular Devices, Sunnyvale, California). Concentrations of the reagents at t0 were 333 nM for PDI, 2.4 µM for DiE-GSSG, 33 µM for DteT, and 0.1 nM to 100 µM for the various molecules under study; except ammonium sulphate and sodium chloride (0.5 M).
Results
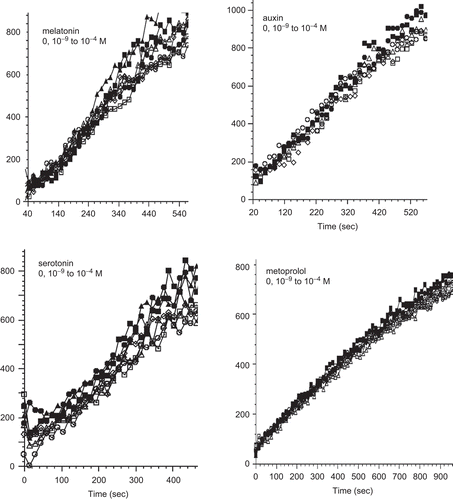
In the course of screening for molecules affecting PDI reductase activity using DiE-GSSG as substrate we found a number of molecules without any effect including metoprolol, serotonin, melatonin, and auxin ().
Figure 1. Kinetics of di-eosin oxidized glutathione reduction into fluorescent E-GSH catalyzed by protein disulphide isomerase in the presence of various concentrations (0–10−4 M) of melatonin, auxin, serotonin or metoprolol.
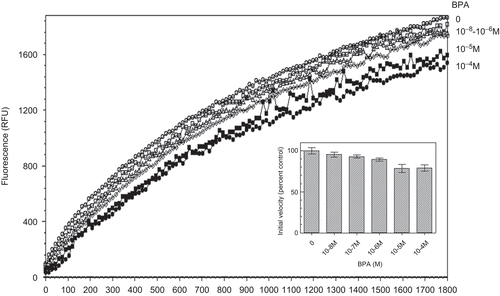
Since potent oestrogenic molecules were found to inhibit PDI reductase activity, we looked for a possible effect of the weak oestrogen BPA. shows that BPA only exerted a slight inhibitory effect on PDI reductase activity in the 10−6–10−5 M range.
Figure 2. Kinetics of di-eosin oxidized glutathione reduction into fluorescent E-GSH catalyzed by protein disulphide isomerase in the presence of various concentrations (0–10−4 M) of bisphenol A. BPA, bisphenol A; RFU, relative fluorescence unit.
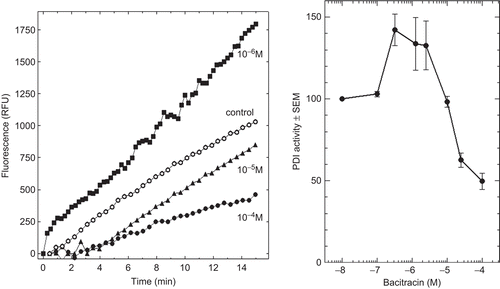
In order to get further information on PDI-inhibiting molecules, we included BAC that has been known for a long time as an inhibitor of PDI activity. In fact, shows that BAC exerted a biphasic effect on PDI reductase activity. It potentiated PDI activity at around 10−6 M final concentration and inhibited it at around 10−4 M final concentration.
Figure 3. Effect of increasing concentrations of bacitracin (BAC) on protein disulphide isomerase (PDI) reductase activity. Left panel: kinetics of di-eosin oxidized glutathione reduction into fluorescent E-GSH catalyzed by PDI in the presence of various concentrations (0–10−4 M) of BAC. Right panel: initial velocity of PDI reducing activity as a function of BAC concentration relative to the velocity in the absence of bacitracin taken as control (n = 3). RFU, relative fluorescence unit.
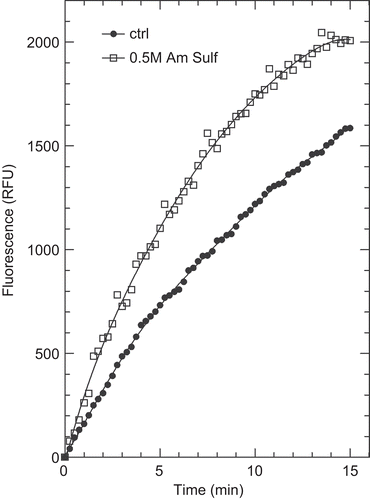
Interestingly, we observed two new molecules in addition to 19-NT and MPACitation5 exhibiting stimulatory effect on PDI reductase activity. Indeed shows that fluoxetin exerted such a potentiating effect but only in the millimolar range. shows that 0.5 M ammonium sulphate also potentiates PDI reductase activity whereas 0.5 M sodium chloride was without any effect (not shown). We also checked that the increase in fluorescence due to ammonium sulphate was indeed due to a higher production of E-GSH and not to higher fluorescence efficiency.
Figure 4. Kinetics of di-eosin oxidized glutathione reduction into fluorescent E-GSH catalyzed by protein disulphide isomerase in the presence of various concentrations (0, 10−4, 10−3 M) of fluoxetine (FLX). The figure also shows the effect of 10% ethanol alone as also present with 10−3 M FLX.
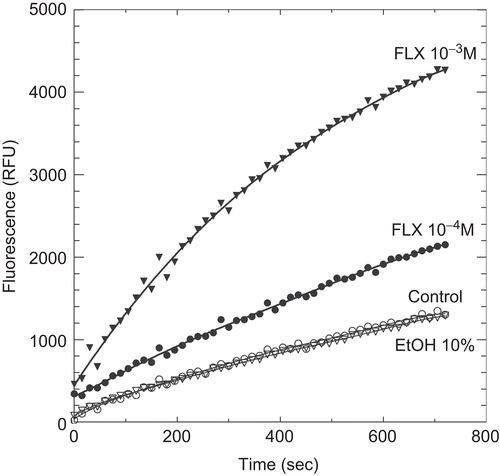
Figure 5. Kinetics of di-eosin oxidized glutathione reduction into fluorescent E-GSH catalyzed by protein disulphide isomerase in the presence or absence of 0.5 M ammonium sulphate. RFU, relative fluorescence unit.
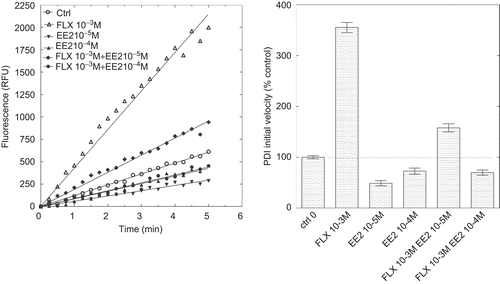
In order to get a better understanding of the mechanisms of ligands inhibitory and stimulatory effects on PDI reductase activity, we analyzed PDI activity in the simultaneous presence of both potentiating and inhibitory ligands. shows the inhibition of PDI activity by increasing doses of DES in the presence of a potentiating dose of 19-NT whereas shows the inhibitory effect of EE2 in the presence of a potentiating concentration of FLX.
Figure 6. Inhibition of protein disulphide isomerase (PDI) reductase activity by diethylstilbestrol (DES) in the presence of a potentiating concentration of 19-nortestosterone (19-NT). Left panel: kinetics of di-eosin oxidized glutathione reduction into fluorescent E-GSH catalyzed by PDI in the presence of 10−5 M of either DES or 19-NT or in the presence of 10−5 M 19-NT together with various concentrations (10−9, 10−7, 10−5 M) of DES. Right panel: initial velocity of PDI reducing activity in the presence of either DES or 19-NT alone or in combination (data from left panel). RFU, relative fluorescence unit.
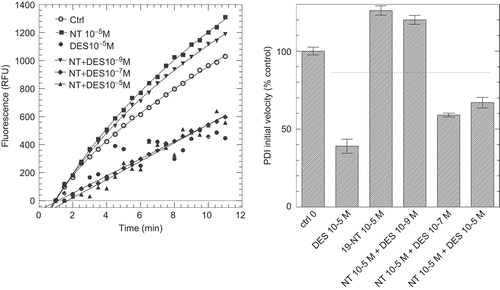
Figure 7. Inhibition of protein disulphide isomerase (PDI) reductase activity by ethynylestradiol (EE2) in the presence of a potentiating concentration of fluoxetine (FLX). Left panel: kinetics of di-eosin oxidized glutathione reduction into fluorescent E-GSH catalyzed by PDI in the presence of 10−3 M FLX or 10−5 or 10−4 M EE2 alone or in the presence of 10−3 M FLX. Right panel: initial velocity of PDI reducing activity in the presence of either FLX or EE2 alone or in combination. RFU, relative fluorescence unit.
Discussion
PDI is a multifunctional enzyme mainly found in the endoplasmic reticulum of eukaryotesCitation7 where its main function is to catalyze the rearrangement (isomerization) of disulphide bridges during folding of membrane and secreted proteins. This activity is of utmost importance as over one-third of all human proteins fold in the endoplasmic reticulumCitation8. The concentration of PDI in the lumen of the endoplasmic reticulum is known to be very highCitation9 and it has been reported to act as a high capacity reservoir for various ligands including hormones such as oestradiol (E2) and thyroxine (T3Citation4,Citation10).
It has been previously shown that all the potent oestrogenic molecules tested (E2, EE2, DES) exhibited an inhibitory effect on PDI isomeraseCitation4,Citation11 and reductaseCitation5 activities. In the present work, we show that the weaker oestrogen BPA also exerts an inhibitory effect on PDI reductase activity. This result is consistent with previous observation that BPA inhibited the chaperone activity of PDI on RNAse renaturationCitation12,Citation13. We found BPA to inhibit only partially (15%) PDI reductase activity and it did so at much higher concentrations (10−5–10−4 M) than the more oestrogenic molecules (10−9–10−8 M). This difference between the inhibitory doses of BPA and those of the potent oestrogenic molecules (E2, EE2, DES) on PDI reductase activity is roughly in the same order of magnitude as their relative oestrogenic activitiesCitation14–21. It can be that, in some way, the binding sites for these different molecules in the oestrogen receptors (ER) and in PDI, respectively are similar enough to explain their similar relative binding affinities for EE2 and BPA, respectively (although this ratio is variable among species and between males and females). Another more exciting possibility relies on the observations that PDI is also present in locations other than the endoplasmic reticulumCitation22 and can directly interact with the ERα and modify its functional propertiesCitation23. A provoking but tenable hypothesis is that the binding of oestrogenic molecules to PDI would play a pivotal role in the stimulation of ER through this direct ERα-PDI interaction.
Since serotonin, melatonin, and auxin are derived from the indolic di-cyclic amino-acid tryptophan, we suspected that they could also exert some inhibitory effect on PDI activity. In fact, we found that they all had no effect on PDI activity at concentrations up to up to 10−4 M (). The β-1 adrenergic receptor antagonists, metoprolol () and atenolol (not shown) also did not exert any effect on PDI activity.
In a previous paperCitation5, we reported that two non-oestrogenic steroids, MPA, and 19-NT potentiated PDI reductase activity. Even with a somewhat limited screening, in the present study we were able to identify a few other molecules also potentiating PDI reductase activity using the recently developed assay with DiE-GSSG as substrateCitation6.
Surprisingly, the well-known inhibitor of PDI activity BACCitation24,Citation25 was found to exert potentiating activity and it did so at lower concentrations (around 10−6 M) than those leading to the expected inhibition (around 10−4 M). BAC is a partly-cyclic polypeptideCitation26 and to our knowledge, there is no clear structural explanation for its mechanism of inhibition of PDI activities. The present data showing a biphasic dose-response effect of BAC on PDI reductase activity might be helpful in future studies to better understand the PDI mechanism of action.
The most efficient potentiating molecule we have identified so far is FLX which is known to act as a selective-serotonin-reuptake inhibitor and is the active component in the antidepressant Prozac™Citation27. FLX was found in our study to greatly increase PDI reductase activity but this effect was observed only at millimolar concentration which is largely higher than its active circulating concentrationsCitation28. The observed augmentation of PDI reductase activity by FLX is therefore not responsible for any of its favourable or unfavourable pharmacological effectsCitation29–33. Nevertheless, the molecular structure of FLX offers an interesting basis for the search of molecules with higher PDI potentiating activities, in order to better understand the molecular mechanism of this enzyme.
Ammonium sulphate (0.5 M) was also found to potentiate PDI reductase activity in contrast to 0.5 M NaCl. It is likely that the mechanism involved is totally different from those implied for 19-NT, BAC, or FLX. The chaotropic/kosmotropic property of ammonium sulphate is well known and greatly influences protein stability and activityCitation34,Citation35, and their crystallization capacityCitation36. It is therefore possible that ammonium sulphate can influence PDI catalytic efficiency by favouring its active conformation as previously shown for glutamate decarboxylaseCitation37 and consistent with the observation that PDI, like thioredoxins and transgtlutaminases, resist detergents and chaotropic agentsCitation38. Another possibility is that ammonium sulphate act by altering water properties and therefore either water-protein interactionsCitation35 or the active concentration of substrate. This latter mechanism would thus be independent of PDI properties.
To our knowledge, this paper and the previous one from our laboratoryCitation5 are the first to describe ligands that potentiate PDI activity. Molecules with such a potentiating effect on PDI activity would be of utmost interest for the treatment of pathologies originating from intracellular protein aggregation. This concerns mainly neurodegenerative diseases such as Alzheimer diseaseCitation39,Citation40 but augmentation of PDI activity would also be expected to have favourable effects in situations where its inhibition by millimolar concentrations of BAC was found detrimental such as in angiogenesisCitation41 or stroke protectionCitation42.
The molecules we have shown so far to exert augmentation of PDI reductase activity exhibit dissimilar structures and it is thus likely that they act through different mechanisms. It will be of interest to broaden the screening for PDI activity-potentiating molecules in order to identify highly potent and possibly synergistic drugs favouring correct folding and disulphide bridges formation in proteins.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Goldberger RF, Epstein CJ, Anfinsen CB. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem 1963;238:628–635.
- Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol 2002;3:836–847.
- Hatahet F, Ruddock LW. Substrate recognition by the protein disulfide isomerases. Febs J 2007;274:5223–5234.
- Primm TP, Gilbert HF. Hormone binding by protein disulfide isomerase, a high capacity hormone reservoir of the endoplasmic reticulum. J Biol Chem 2001;276:281–286.
- Klett D, Cahoreau C, Villeret M, Combarnous Y. Effect of pharmaceutical potential endocrine disruptor compounds on protein disulfide isomerase reductase activity using di-eosin-oxidized-glutathione. Plos one 2010;5:e9507.
- Raturi A, Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic Biol Med 2007;43:62–70.
- Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem Sci 2006;31:455–464.
- Chen Y, Zhang Y, Yin Y, Gao G, Li S, Jiang Y et al. SPD–a web-based secreted protein database. Nucleic Acids Res 2005;33:D169–D173.
- Abell BA, Brown DT. Sindbis virus membrane fusion is mediated by reduction of glycoprotein disulfide bridges at the cell surface. J Virol 1993;67:5496–5501.
- Gilbert HF, Kruzel ML, Lyles MM, Harper JW. Expression and purification of recombinant rat protein disulfide isomerase from Escherichia coli. Protein Expr Purif 1991;2:194–198.
- Tsibris JC, Hunt LT, Ballejo G, Barker WC, Toney LJ, Spellacy WN. Selective inhibition of protein disulfide isomerase by estrogens. J Biol Chem 1989;264:13967–13970.
- Hiroi T, Okada K, Imaoka S, Osada M, Funae Y. Bisphenol A binds to protein disulfide isomerase and inhibits its enzymatic and hormone-binding activities. Endocrinology 2006;147:2773–2780.
- Hashimoto S, Okada K, Imaoka S. Interaction between bisphenol derivatives and protein disulphide isomerase (PDI) and inhibition of PDI functions: requirement of chemical structure for binding to PDI. J Biochem 2008;144:335–342.
- Ashby J, Odum J. Gene expression changes in the immature rat uterus: effects of uterotrophic and sub-uterotrophic doses of bisphenol A. Toxicol Sci 2004;82:458–467.
- Schmidt S, Degen GH, Seibel J, Hertrampf T, Vollmer G, Diel P. Hormonal activity of combinations of genistein, bisphenol A and 17beta-estradiol in the female Wistar rat. Arch Toxicol 2006;80:839–845.
- Takagi H, Shibutani M, Masutomi N, Uneyama C, Takahashi N, Mitsumori K et al. Lack of maternal dietary exposure effects of bisphenol A and nonylphenol during the critical period for brain sexual differentiation on the reproductive/endocrine systems in later life. Arch Toxicol 2004;78:97–105.
- Sohoni P, Tyler CR, Hurd K, Caunter J, Hetheridge M, Williams T et al. Reproductive effects of long-term exposure to Bisphenol A in the fathead minnow (Pimephales promelas). Environ Sci Technol 2001;35:2917–2925.
- Stoker C, Rey F, Rodriguez H, Ramos JG, Sirosky P, Larriera A et al. Sex reversal effects on Caiman latirostris exposed to environmentally relevant doses of the xenoestrogen bisphenol A. Gen Comp Endocrinol 2003;133:287–296.
- Yamasaki K, Sawaki M, Noda S, Imatanaka N, Takatsuki M. Subacute oral toxicity study of ethynylestradiol and bisphenol A, based on the draft protocol for the “Enhanced OECD Test Guideline no.407”. Arch Toxicol 2002;76:65–74.
- Trudeau VL, Turque N, Le Mé vel S, Alliot C, Gallant N, Coen L et al. Assessment of estrogenic endocrine-disrupting chemical actions in the brain using in vivo somatic gene transfer. Environ Health Perspect 2005;113:329–334.
- Ceccarelli I, Della Seta D, Fiorenzani P, Farabollini F, Aloisi AM. Estrogenic chemicals at puberty change ERalpha in the hypothalamus of male and female rats. Neurotoxicol Teratol 2007;29:108–115.
- Turano C, Coppari S, Altieri F, Ferraro A. Proteins of the PDI family: unpredicted non-ER locations and functions. J Cell Physiol 2002;193:154–163.
- Schultz-Norton JR, McDonald WH, Yates JR, Nardulli AM. Protein disulfide isomerase serves as a molecular chaperone to maintain estrogen receptor alpha structure and function. Mol Endocrinol 2006;20:1982–1995.
- Roth RA. Bacitracin: an inhibitor of the insulin degrading activity of glutathione-insulin transhydrogenase. Biochem Biophys Res Commun 1981;98:431–438.
- Mandel R, Ryser HJ, Ghani F, Wu M, Peak D. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc Natl Acad Sci USA 1993;90:4112–4116.
- Stone KJ, Strominger JL. Mechanism of action of bacitracin: complexation with metal ion and C 55 -isoprenyl pyrophosphate. Proc Natl Acad Sci USA 1971;68:3223–3227.
- Wong DT, Perry KW, Bymaster FP. Case history: the discovery of fluoxetine hydrochloride (Prozac). Nat Rev Drug Discov 2005;4:764–774.
- Pérez V, Puiigdemont D, Gilaberte I, Alvarez E, Artigas F. Augmentation of fluoxetine’s antidepressant action by pindolol: analysis of clinical, pharmacokinetic, and methodologic factors. J Clin Psychopharmacol 2001;21:36–45.
- Jang DP, Lee SH, Park CW, Lee SY, Kim YB, Cho ZH. Effects of fluoxetine on the rat brain in the forced swimming test: a [F-18]FDG micro-PET imaging study. Neurosci Lett 2009;451:60–64.
- Lister A, Regan C, Van Zwol J, Van Der Kraak G. Inhibition of egg production in zebrafish by fluoxetine and municipal effluents: a mechanistic evaluation. Aquat Toxicol 2009;95:320–329.
- Lee CS, Kim YJ, Jang ER, Kim W, Myung SC. Fluoxetine induces apoptosis in ovarian carcinoma cell line OVCAR-3 through reactive oxygen species-dependent activation of nuclear factor-kappaB. Basic Clin Pharmacol Toxicol 2010;106:446–453.
- Lee LJ. Neonatal fluoxetine exposure affects the neuronal structure in the somatosensory cortex and somatosensory-related behaviors in adolescent rats. Neurotox Res 2009;15:212–223.
- Lim CM, Kim SW, Park JY, Kim C, Yoon SH, Lee JK. Fluoxetine affords robust neuroprotection in the postischemic brain via its anti-inflammatory effect. J Neurosci Res 2009;87:1037–1045.
- Yang Z, Liu XJ, Chen C, Halling PJ. Hofmeister effects on activity and stability of alkaline phosphatase. Biochim Biophys Acta 2010;1804:821–828.
- Yang Z. Hofmeister effects: an explanation for the impact of ionic liquids on biocatalysis. J Biotechnol 2009;144:12–22.
- Dumetz AC, Lewus RA, Lenhoff AM, Kaler EW. Effects of ammonium sulfate and sodium chloride concentration on PEG/protein liquid-liquid phase separation. Langmuir 2008;24:10345–10351.
- Hiraga K, Ueno Y, Oda K. Glutamate decarboxylase from Lactobacillus brevis: activation by ammonium sulfate. Biosci Biotechnol Biochem 2008;72:1299–1306.
- Rao RU, Mehta K. Transglutaminases, thioredoxins and protein disulphide isomerase: diverse enzymes with a common goal of cross-linking proteins in lower organisms. Indian J Exp Biol 2004;42:235–243.
- Kim HT, Russell RL, Raina AK, Harris PL, Siedlak SL, Zhu X et al. Protein disulfide isomerase in Alzheimer disease. Antioxid Redox Signal 2000;2:485–489.
- Dinoto L, Deture MA, Purich DL. Structural insights into Alzheimer filament assembly pathways based on site-directed mutagenesis and S-glutathionylation of three-repeat neuronal Tau protein. Microsc Res Tech 2005;67:156–163.
- Tian F, Zhou X, Wikström J, Karlsson H, Sjöland H, Gan LM et al. Protein disulfide isomerase increases in myocardial endothelial cells in mice exposed to chronic hypoxia: a stimulatory role in exposure after angiogenesis. Am J Physiol Heart Circ Physiol 2009;297:H1078–H1086.
- Descamps E, Petrault-Laprais M, Maurois P, Pages N, Bac P, Bordet R et al. Experimental stroke protection induced by 4-hydroxybenzyl alcohol is cancelled by bacitracin. Neurosci Res 2009;64:137–142.
