Abstract
A number of isolates from different ecosystems were screened for their ability to inhibit tyrosinase resulting in the selection of isolate CFR 101, which showed an inhibition of 72%. The metabolites present in the crude extract of the selected isolate was profiled through high-performance liquid chromatography (HPLC) before the enzyme inhibition assay to reveal a 66% decrease in area of the peak at room temperature for 13.9 min, after the assay. Upon purification, this peak was identified as kojic acid, a known inhibitor of tyrosinase. This unique technique of combining a reaction assay mixture with HPLC profile wherein inhibitors can be rapidly pinpointed in crude extracts addresses the drawback of rapid chemical high-throughput screening (HTS) systems, which is limited to the chemical nature of metabolites without any evidence of their biological activities.
Keywords::
Introduction
Integrating chromatography with different analytical techniques provides a powerful tool for screening of new molecules from nature. As high-throughput screening (HTS) gained popularity, it became important to develop methods to quickly eliminate known compoundsCitation1. One such tool was the use of liquid chromatography (LC)–UV–masss spectrometry (MS) to profile fungal metabolites and mycotoxinsCitation2, which helped create a database of 474 metabolites from microfungi. Another example of integration is that of chromatography and MSCitation3. The biggest drawback of these techniques is that they focus on the physical presence of metabolites and do not show any evidence of their biological activities, such as enzyme inhibition, antibiotic or antimicrobial effects, etc.
Almost all the assays for the detection and quantification of enzyme inhibitors are based on either colorimetricCitation4 or amperometricCitation5 methods. A simple method of incubation of an enzyme, inhibitor and substrate offers the scope for any number of permutations and combinations in free and immobilised supports to detect a potent inhibitor in a mixture. For example, immobilisation of proteases on a solid support has been considered a potentially powerful procedure as it allows effective protease concentrations, due to reduced autolysis of productsCitation6. Several groups have explored the application of immobilised proteases on large-scale proteomic researchCitation7. The work described here introduces a unique technique of combining a biological assay with an analytical technique wherein inhibitors can be rapidly pinpointed in crude extracts through an immobilised-enzyme-catalysed reaction assay and profiled through high-performance liquid chromatography (HPLC).
Materials and methods
Materials
Glass beads with average diameters of 0.3 mm, 3-aminopropyl-triethoxysilane (APTES), tyrosinase enzyme (polyphenol oxidase), l-DOPA, bovine serum albumin was obtained from Sigma Aldrich. Coomassie Brilliant Blue G-250 was obtained from Merck. All other chemicals were of analytical grade and were purchased from Merck AG. Double distilled water was used throughout.
Organisms
One g of air dried soil samples collected from various ecosystems were serially diluted in sterile saline and plated on potato dextrose agar (PDA). Upon growth, individual colonies were transferred to other plates successively until the isolation of pure cultures. These fungal isolates were maintained on PDA slants and refrigerated at 7°C and the cultures were sub-cultured every fortnight.
Fermentation and extraction of crude extracts
Cultures were inoculated onto 500-mL Erlenmeyer flasks containing 150 mL potato dextrose broth and kept for incubation at 200 r.p.m. at 30°C for 7 days. At the end of fermentation, 150 mL of ethyl acetate was added to the fermented broth and stirred for an hour at room temperature. The solvent was separated and evaporated using a rotator vacuum distilling apparatus. The resulting crude extract was taken as a source of enzyme inhibitor.
Glass bead immobilisation
The method of MunneckeCitation8 was followed. Briefly, the glass beads were treated with methanol for 2 h at 45°C, followed by 5% nitric acid for 2 h at 80°C, and washed with distilled water and dried. The beads were then silanised with 25 mL of 3% (v/v) solution of γ-APTES in acetone at room temperature for 3 h with intermittent shaking. The amino functionalised glass beads were removed from solution by filtration and kept in the oven at 110°C overnight to achieve complete formation of Si–O–Si bonds. A volume of 25 mL of 2.5% glutaraldehyde in sodium phosphate buffer of pH 6.0, 50 mM was added to silanised golden yellow glass beads for 2 h at 30°C and washed several times with distilled water.
A 1000 U of tyrosinase enzyme in 2 mL 100 mM phosphate buffer (pH 6.8) was added to the silanised glass beads and incubated for 3 h at 30 °C, followed by addition of 50 mg of sodium cyanoborohydride and incubated at 4°C for 19 h. After washing the enzyme-immobilised glass beads several times with distilled water and then with phosphate buffer (pH 6.8 ), 10 mL ethanolamine solution 0.124 M was added for 3 h at 4°C. The beads were again washed and stored in buffer at 4°C. Before the start of the experiments, the enzyme protein concentration was measured using Bradford’s method and the amount of bound protein per weight of support was calculated.
Tyrosinase assays
The enzyme assay was carried out according to the method of Duckworth and ColemanCitation9 using purified mushroom tyrosinase with a specific activity of 3300 U/mg protein. A typical enzyme assay comprised 10 µL of enzyme; 10 µL of crude extract (inhibitor) dissolved in dimethyl sulfoxide (DMSO) and made up to 1 mL by 100 mM phosphate buffer (pH 6.8). In the control assay, same volume of DMSO was used instead of the inhibitor. The enzyme reaction was initiated by the addition of 35 µL substrate (6.3 mM DOPA) and the increase in absorbance at 475 nm was recorded at the end of 3 min. The relative activity was expressed as the percentage ratio of enzyme activity in the presence of inhibitor with enzyme activity in the absence of enzyme inhibitors at the end of 3 min of enzyme reaction.
In the case of immobilised enzyme, five beads corresponding to 1.644 µg protein, was dispersed in 400 µL of 100 mM phosphate buffer to form a uniform suspension and incubated with 10 μL of crude extract for 5 min. The incubated solution was removed and beads washed with buffer. A 10 μL of the substrate (6.3 mM DOPA) was added to the beads and incubated for 15 min, allowing for product formation The reaction was monitored in Quartz cuvette with a 1-cm light path with 10 μL of the solution from tube and 1mL of buffer. The enzyme reaction was monitored by the increase in absorbance spectrum of 800–200 nm and compared with the enzymatic reaction without inhibitor (control). The percentage of inhibition was calculated. The presence of inhibitor attached to enzyme was also confirmed by acid washing with dilute HCl.
HPLC analysis
HPLC was carried out in Shimadzu LC-10 with a diode array system. A stock of 40 μg/mL crude extract was prepared in methanol and from this 20 μL was injected in to the reverse phase C18 column (4.6 × 250 mm). Elution was achieved with isocratic phosphate buffer: methanol (99:1) pH 2.5.at a flow rate of 0.9 mL/min. The detector used was photodiode array detector the detection wavelength was 269 and 280 nm.
Purification of the inhibitor
The crude extract was coated on 0.5 g silica gel and applied to a column (10 × 500mm) of silica gel (60–120 mesh) that had been dried at 150°C overnight and packed in hexane with flow rate 1 mL/min. Elution of crude was carried out using hexane, chloroform, ethyl acetate and methanol. The fractions were analysed for activity by enzyme assay. The active fractions were pooled up and further purified by size exclusion chromatography (Sephadex LH-60). The purified compound was checked for inhibition.
Identification of the inhibitor
The structure of the compound isolated from CFR 101 was determined by spectroscopic analysis. The nuclear magnetic resonance (NMR) spectra were recorded on Bruker, DRX-400 MHz instrument at 20°C. About 10 mg of the solid sample dissolved in DMSO-d6 was used for recording the spectra. Infrared (IR) spectra were obtained in KBr pellets with a Bio-Rad FTS-135 spectrophotometer (Bio-Rad, Richmond, CA). Electro spray ionization MS (ESIMS) data were taken on a Waters-3000 mass spectrometer.
Kinetic analysis of tyrosinase inhibition
The reaction mixture consisted of four different concentrations of l-3,4-dihydroxyphenylalanine (l-DOPA; 1.5–12.6 mM) as a substrate and mushroom tyrosinase in 0.1 M sodium phosphate buffer. Different concentrations of kojic acid were added to the reaction mixture, respectively. Ki and mode of inhibition of the inhibitor was determined by Dixon plot.
Results
Screening of fungal isolates
A total of 50 fungal isolates were selected and each was subjected to fermentation on potato dextrose broth. The crude fermented extracts were then screened for their ability to inhibit tyrosinase by conventional free-enzyme assay. The results are shown in . Among the 50 isolates screened for tyrosinase inhibitor, the CFR 101 was found to be active (72%) in comparison with the rest.
Table 1. Results of the screening of fungal isolates against free enzyme tyrosinase.
HPLC of crude extract before assay
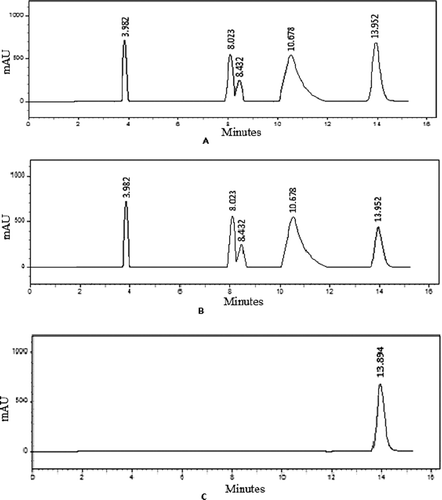
The HPLC profile of the crude extract of isolate CFR 101 was done using various mobile phase systems in order to obtain good separation of peaks. phosphate buffer and methanol (99:1) gave a good separation and were selected. The profile is shown in .
HPLC of crude extract after assay
The crude was added to the immobilised enzyme and incubated for 5 min and then the supernatant was injected to HPLC. The percentage reduction in all the peak areas were calculated (). It was observed that while the reduction in the peak areas from 3.9 to 10.6 min was marginal, there was a 66% reduction in the peak area at 13.9 min (). To confirm the presence of the inhibitor at 13.9 min, the immobilised enzyme after the incubation was treated with methanol. An HPLC profile of the methanolic wash confirmed the presence of the inhibitor from isolate CFR 101 at 13.9 min.
Table 2. Reduction in peak area of metabolites of CFR 101 before and after the enzyme assay.
Conventional purification of inhibitor
Three litres of fermented CFR 101 extracted with ethyl acetate yielded 1.5 g of crude extract. The purification was carried out in two stages of column chromatography. The crude extract was passed through silica gel (60–120) column where different fractions obtained with hexane, chloroform and ethyl acetate were checked for inhibition and spectrum scan. Three active fractions of 100 mL each were collected at an eluent composition of hexane:chloroform (1:1 v/v), hexane:chloroform (0.3:0.7 v/v) and chloroform, respectively The active fractions showing inhibition were pooled again and loaded onto the Sephadex LH-60 gel filtration column. In this second stage of chromatography, the partially purified fractions were eluted with methanol to get a pure white crystalline powder.
Structure elucidation of the inhibitor
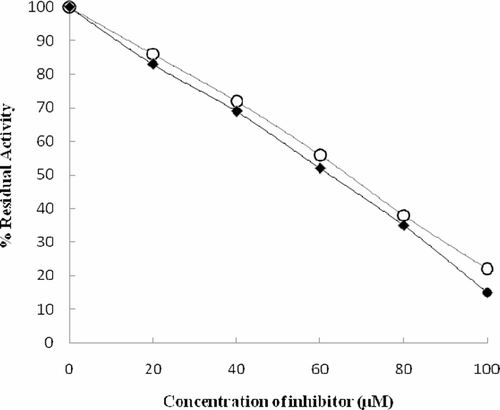
The structure of the compound was elucidated by performing NMR, mass and IR spectroscopy and was identified as kojic acid. The inhibitor from CFR101 was confirmed to be kojic acid when compared with standard kojic acid wherein both showed the same retention time (13.952 min) and m/z (143.1) by HPLC and MS, respectively. Similarly, A dose dependency of tyrosinase inhibition by isolated and analytical grade kojic acid reveals that their IC50 values were 69 and 63 µM, respectively ().The slightly higher IC50 value of the isolated kojic acid is perhaps due to the presence of small quantities of impurities.
Stability and reuse of immobilised bead
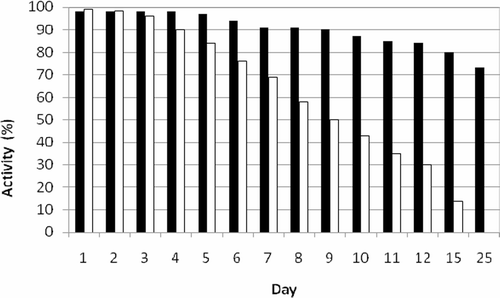
The stability of the immobilised enzyme was studied where in the immobilised enzyme and free enzymes were stored at 4°C in phosphate buffer (100 mM pH 6.8); the activity measurements were carried out for a period of 25 days. The free enzyme lost all its activity within 25 days. The immobilised tyrosinase was stable up to 5 days and lost only 25% of activity in 25 days ().
Figure 3. Stability of immobilised and free tyrosinase. Free tyrosinase (open bar), immobilised tyrosinase (closed bar).
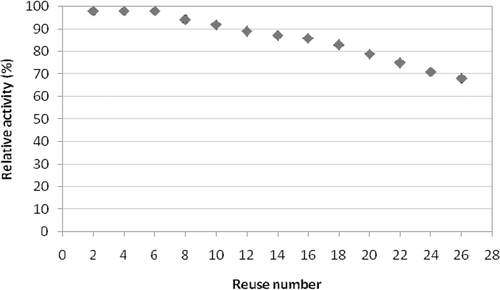
The reuse of the immobilised enzymes for screening for inhibitors was also evaluated. The immobilised enzyme was washed with buffer and re-incubated with the same freshly prepared crude extract and the activity was monitored. This was repeated 26 times. Even though there was consistent inhibition, the immobilised tyrosinase retained 90% of its original activity up to 7 reuses and lost 20% of original activity up to 26th reuse ().
Discussion
The implication of enzymes in various disease states has focused attention on the search for enzyme inhibitors among the various sources of natural productsCitation10, leading to a flurry of discoveries of new enzyme inhibitors.
Use of tyrosinase inhibitors is becoming increasingly important in the cosmetic and medicinal industries due to their preventive effect on pigmentation disordersCitation11. Tyrosinase inhibitors may result in a reduction in melanin biosynthesis and are used in cosmetic products for hyper-pigmentation-related concerns, including the formation of freckles. Tyrosinase and its inhibitors may also be targets for developing medicines to treat hypo-pigmentation-related problems, such as albinism and piebaldismCitation12.
In continuation of our discovery program on new enzyme inhibitors from microbial sourcesCitation13–15, the soil from various ecosystems was plated on PDA. Out of the 50 isolates screened for the tyrosinase inhibition activity, a primary hit CFR 101 with 72% inhibition was selected for profiling using our PRofiling of Enzyme-Metabolite Assay (PREMA)-HPLC method. An HPLC profile of crude supernatant before incubation with the immobilised enzyme, crude supernatant got after incubation with the immobilised enzyme and the compound obtained after the immobilised enzyme was washed with 10% methanol and phosphate buffer and methanol 99:1 as mobile phase (). The percentage reduction in the peak areas of crude supernatant before and after incubation was marginal with the retention time of 3.9–10.6 min but a significant reduction of 66% at 13.9 min indicated the presence of a compound in the crude, a part of which got bound with the enzyme during the incubation period (). This was confirmed when a single peak was obtained at 13.9 min with compound obtained after washing the immobilised enzyme with 10%methanol. After the identification of the inhibitory compound, purification of the compound from the crude was done by adsorption and gel permeation chromatography in tandem to get a pure white crystalline powder.
The structure of the compound was elucidated by performing NMR, mass and IR spectroscopy and was identified as kojic acid with molecular formula of C6H6O4. Further confirmation about the identity of the compound was done by the activity profiling of the compound, in which the compound has shown an IC50 value of 69 µM, where the standard kojic acid has shown an IC50 value of 63 µM, the slightly higher IC50 value of the isolated compound is due to the presence of mere amount of impurities ().
One of the most important parameters to be considered in enzyme immobilisation is storage stability. In general, if an enzyme is in solution, it is not stable during storage, and the activity is gradually reduced. The effect of storage conditions on the activity of the immobilised enzyme is an important aspect to ensure that a long shelf life is possible. The decline of enzyme activity is essentially identical for all storage condition testedCitation16. From the stability studies carried out for both free and immobilised enzymes stored in phosphate buffer at 4°C, it was determined that the free enzyme lost all its activity within 25 days and the immobilised tyrosinase was stable up to 5 days and lost only 25% of activity in 25 days ().
The reusability of the immobilised enzyme carries the same importance as that of stability of the immobilised enzyme. The reusability of the immobilised tyrosinase was studied monitoring the activity of the enzyme after each use. this was done for 26 times, by which the immobilised tyrosinase retained 90% of its original activity up to 7 reuses and lost 20% of original activity up to 26th reuse ().
Conclusions
One of the tough challenges of researchers involved in natural product discovery is to arrive at biologically active purified compounds from crude extracts containing hundreds of known and unknown compounds, in the least amount of time with minimum unit operations of purification. Even though a lot of work going on in the area of metabolomics and HTS, much of the progress is seen in a combination of physical and analytical techniques such as gas chromatography–MS, LC–MS or LC–UV–MS, LC–NMR, etcCitation17–19. Central to the discovery of new bioactive molecules is the biological assay. Integrating a biological activity or assay with purification has been successful in antibiotics research with the development of bioautographs, which combines antimicrobial bioassay with (paper) chromatographic purification in a single stepCitation20,Citation21. In this article, even though a known molecule, kojic acid, was isolated as a tyrosinase inhibitor, the technique of combining a reaction assay mixture with HPLC is novel and can be adapted to rapidly screen any target enzyme, immobilised on a glass support, for inhibitors from crude mixtures. After incubation, the identification of the peak at a particular retention time decreases significantly, also helps in developing strategies for its purification through preparative HPLC, thus saving time and down streaming costs. Even though this is a novel approach to rapidly screen for inhibitors from crude mixtures and can be easily scaled up to handle HTS samples, further work on assessing this method’s performance in presence of tight-binding inhibitors is being done along with strategies to integrate de-replication and sift known inhibitors from new ones.
Acknowledgements
The funding for this work from the 11th Five Year Plan Network Project on “Exploitation of India’s rich microbial diversity” is gratefully acknowledged. The assistance of Mr. Dasagrandhi Chakradhar in the early part of this work is acknowledged.
Declaration of interest
The funding for this work from the 11th Five Year Plan Network Project on “Exploitation of India’s rich microbial diversity” is gratefully acknowledged.
References
- Schuster E, Dunn-Coleman N, Frisvad JC, Van Dijck PW. On the safety of Aspergillus niger—a review. Appl Microbiol Biotechnol 2002, 59, 426–435.
- Fog NK, Jorn S. Fungal metabolite screening: Database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography–UV–mass spectrometry methodology. J Chromatogr A 2002;1002:111–136.
- Strege MA. High-performance liquid chromatographic–electrospray ionization mass spectrometric analyses for the integration of natural products with modern high-throughput screening. J Chromatogr A 1999;725:67–68.
- Serra CP, Côrtes SF, Lombardi JA, Braga de Oliveira A, Braga FC. Validation of a colorimetric assay for the in vitro screening of inhibitors of angiotensin-converting enzyme (ACE) from plant extracts. Phytomedicine 2005;12:424–432.
- Jeanty G, Wojciechowska A, Marty JL, Trojanowicz M. Flow-injection amperometric determination of pesticides on the basis of their inhibition of immobilized acetylcholinesterases of different origin. Anal Bioanal Chem 2002;373:691–695.
- Parrado J, Millan F, Bautista J. Kerase immobilization by covalent attachment to porous-glass. Process Biochem 1995;30:735–741.
- Anwar A, Qader SAU, Raiz A, Iqbal S, Azhar A. Calcium alginate: A support material for immobilization of proteases from newly isolated strain of Bacillus subtilis KIBGE-HAS. World Appl Sci J 2009;10:1281–1286.
- Munnecke DM. Hydrolysis of organophosphate insecticides by an immobilized-enzyme system. Biotechnol Bioeng 1979;21:2247–2261.
- Duckworth HW, Coleman JE. Physicochemical and kinetic properties of mushroom tyrosinase. J Biol Chem 1970;245:1613–1625.
- Umezawa H. Low-molecular-weight enzyme inhibitors of microbial origin. Annu Rev Microbiol 1982;36:75–99.
- Chang TS. An updated review of tyrosinase inhibitors. Int J Mol Sci 2009;10:2440–2475.
- Kim YJ, Uyama H. Tyrosinase inhibitors from natural and synthetic sources: structure, inhibition mechanism and perspective for the future. Cell Mol Life Sci 2005;62:1707–1723.
- Sekhar Rao KC, Divakar S, Karanth NG, Sattur AP. 14-(2′,3′,5′-Trihydroxyphenyl)tetradecan-2-ol, a novel acetylcholinesterase inhibitor from Chrysosporium sp. J Antibiot 2001;54:848–849.
- Rao KC, Divakar S, Babu KN, Rao AG, Karanth NG, Sattur AP. Nigerloxin, a novel inhibitor of aldose reductase and lipoxygenase with free radical scavenging activity from Aspergillus niger CFR-W-105. J Antibiot 2002;55:789–793.
- Chidananda C, Rao LJ, Sattur AP. Sclerotiorin, from Penicillium frequentans, a potent inhibitor of aldose reductase. Biotechnol Lett 2006;28:1633–1636.
- Leng HL, Douglas GM, Gordon AH. Hydrolysis of starch particles using immobilized barley α-amylase. Biochem Engg J 2003;13:53–62.
- Hu Z-J, Roddy TP, Ho P-I, Horvath CR, Vickers C, Stout S, Hubbard B, Wang YK, Hill WA, Bojanic D. Assay development and screening of human dGAt1 inhibitors with an LC/MS-based assay: Application of mass spectrometry for large-scale primary screening. J Biomol Screen 2010 (DOI: 10.1177/1087057110370210).
- Liu X, Ashforth E, Ren B, Song F, Dai H, Liu M, Wang J, Xie Q, Zhang L. Bioprospecting microbial natural product libraries from the marine environment for drug discovery. J Antibiot 2010;63:415–422.
- Yu F, Liang K, Zou H, Lei X. Progress on the screening and analysis of bioactive compounds in traditional Chinese medicines by biological fingerprinting analysis. Comb Chem High Throughput Screening 2010;13:855–868.
- Annis SL, Velasquez L, Xu H, Hammerschmidt R, Linz J, Trail F. Novel procedure for identification of compounds inhibitory to transcription of genes involved in mycotoxin biosynthesis. J Agric Food Chem 2000;48:4656–4660.
- Müller MC, Dausend C, Weins C, Frimmel FH. New bioautographic screening method for the detection of estrogenic compounds. Chromatographia 2004;60:207–211.