Abstract
In the present investigation synthesis of some novel 1-(2-(1H-benzimidazol-2-yl)phenyl)-3-chloro-4-(Un/substitutedphenyl)azetidin-2-one (3a-3h) antibacterial are reported. Structures of synthesized compounds were confirmed by spectral techniques (IR, Mass, 1H-NMR) All reactions were monitored with analytical thin layer chromatography. Synthesized compounds were docked in to the active site of enzyme transpeptidase. Compounds 3a, 3b, 3d and 3g were found to have good affinity for transpeptidase with potent antibacterial activity. A good correlation is found between in silico docking analysis and in vitro antibacterial activity.
Keywords::
Introduction
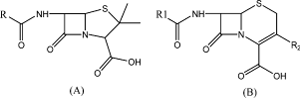
β-lactam antibiotics are a broad class of agents that include penicillin (A) derivatives (penams), cephalosporins (B) (cephems), monobactams, and carbapenems. These agents contain a β-lactam (cyclic amide) nucleus their molecular structureCitation1.
β-Lactam antibiotics are bactericidal, and act by inhibiting the synthesis of the peptidoglycan layer of bacterial cell walls. The peptidoglycan layer is important for cell wall structural integrity, especially in Gram-positive organismsCitation2. The final transpeptidation step in the synthesis of the peptidoglycan is facilitated by transpeptidases known as penicillin-binding proteins (PBPs). The β-lactam nucleus of the molecule irreversibly binds to (acylates) the Ser403 residue of the PBP active site. This irreversible inhibition of the PBPs prevents the final cross linking (transpeptidation) of the nascent peptidoglycan layer, disrupting cell wall synthesisCitation3.
Benzimidazoles and their analogs are well known biologically active N-containing heterocyclesCitation4. Specifically, the 2-substituted analogs of benzimidazoles are known to be potent biologically active compoundsCitation5. Extensive biochemical and pharmacological studies have confirmed that benzimidazole molecules are effective against various strains of microorganismsCitation6,Citation7. Owing to the immense importance as antimicrobial agents exhibited by benzimidazoles, efforts have been made from time to time to generate libraries of these compounds and screened them for antimicrobial activity.
Monocyclic β-lactams have attracted much attention owing to their important pharmacological propertiesCitation8. Some monocyclic β-lactams containing the N-heterocyclic moieties like benzimidazole in the skeleton have shown good antibacterial activityCitation9. The high synthetic utility and pharmacological importance of β-lactam family, medicinal chemist has synthesized many synthetic and semi synthetic potent β-lactams. In the present work, synthesis of some novel 1-(2-(1H-benzimidazol-2-yl)-phenyl)-3-chloro-4-[(un)/substituted phenyl]-azetidin-2-one antibacterial and in silico molecular docking analysis of synthesized scaffold in to the active site of enzyme transpeptidase is reported. In continuation to our work on structure based drug designCitation10,Citation11 and antibacterial researchCitation12,Citation13 in the present investigation structure based design of novel benzimidazoles based monocyclic β-lactam is described. Design of ligands is carried out by in-silico molecular dockingCitation10 in to the active site of enzyme transpeptidase. Ligand with low binding energy is considered to be having high affinity for the receptor transpeptidase.
Materials and methods
Melting points were determined on a Thomas–Hoover melting point apparatus and are uncorrected. IR spectra were recorded on a Jasco infrared spectrometer in KBr. 1H NMR was recorded in dimethyl sulfoxide-d 6 or CDCl3 solutions using a Brucker Avance II 400 spectrophotometer at a frequency of 400.13 MHz. Chemical shifts are given in ppm relative to tetramethylsilane (TMS) as internal standard. The electrospray mass spectra were recorded on MICROMASS QUATRO II triple quadrapole mass spectrophotometer. All the synthesized compounds were analyzed satisfactorily for C, H and N by Elementar Vario EL III elemental analyzer. Analytical thin layer chromatography (TLC) was performed by using adsorbent silica gel G, Visualilization of the developed chromatogram was performed with iodine vapors. Solvents and reagents obtained from commercial sources were used without purification.
Results and discussion
Chemistry
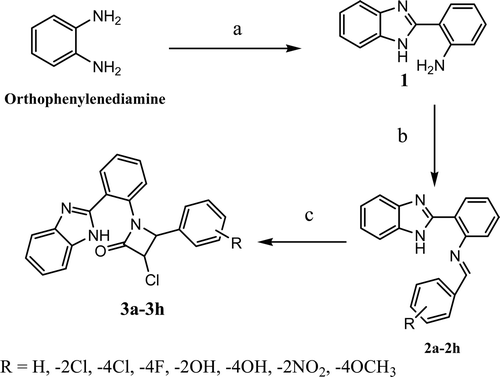
Target compounds 3a-3h are prepared by synthetic protocol described in . 2-(1H-benzimidazol-2-yl)benzenamine (2) was synthesized by refluxing orthophenylenediamine with anthanilic acid in the presence of polyphosphoric acid. Obtained, compound 2 was then refluxed with different aromatic aldehydes in the presence of sulfuric acids to form Schiff bases (2a-2h). Resultant Schiff bases were cyclised in to 2-azetidinones 3a-3h with by refluxing with chloroacetylchloride in the presence of triethylamine. 2-azetidinones can be prepared by simple stirring Schiff base with chroacetylchloride in suitable dimethylsulfoxide for 48 h or by heating on water bath for 4 h. When 2-azetidiones are prepared by heating strictly trans isomers are formedCitation14. All reactions were monitored by analytical thin layer chromatography. The structures of synthesized compounds are confirmed by IR, MS and 1HNMR spectroscopy. Compounds 3a-3h have given the satisfactory elemental analysis by C, H and N. The antibacterial activity of synthesized compound was tested against various Gram positive and Gram negative bacteria stainsCitation12.
In vitro antibacterial activity
The antibacterial activity was tested against various Gram positive and Gram negative bacteria stainsCitation12. Synthesized compounds were screened for their in vitro antimicrobial activity against the standard strains: S. aureus (ATCC 25923), B. subtilis (ATCC 6633), E. coli (ATCC 25922), P. aeruginosa (ATCC 27853). To evaluate the activity of synthesized compounds against bacteria, minimum inhibitory concentrations (MICs) were determined. Ampicillin, the reference β-lactam antibacterial drug was used as positive control. The results are described in the .
Table 1. Antimicrobial activity of synthesized compounds 3a-3h
Molecular modeling studies
β-lactam antibiotics act by inhibiting the synthesis of the peptidoglycan layer of bacterial cell walls. The final transpeptidation step in the synthesis of the peptidoglycan is facilitated by transpeptidases known as penicillin-binding proteins (PBPs). β-Lactam antibiotics are analogues of D-alanyl-D-alanine- the terminal amino acid residues on the precursor NAM/NAG-peptide subunits of the nascent peptidoglycan layer. The structural similarity between β-lactam antibiotics and D-alanyl-D-alanine facilitates their binding to the active site of penicillin-binding proteinsCitation15 (PBPs). The β-lactam nucleus of the molecule irreversibly binds to acylate the Ser403 residue of the PBP active site. This irreversible inhibition of the PBPs prevents the final cross linking (transpeptidation) of the nascent peptidoglycan layer, disrupting cell wall synthesis. Therefore enzyme transpeptidase is an important target for penicillin like antibacterial design. To pre-assess the anti-bacterial behavior of synthesized compounds 3a-3h on a structural basis, automated docking studies were carried out and scoring functions, their binding affinities and orientation of these compounds at the active site of the enzyme transpeptidase were found out. Results are summarized in . The protein–ligand complex was constructed on the basis of the X-ray structure of transpeptidaseCitation15 (PDB code–3PTE). Docking of compounds 3a-3h with known antibacterial agent such as ampicillin was performed using Molecular Design Suite (MDS) v 3.5, into the 3D model of the catalytic site of enzymes transpeptidase .
Figure 1. Best poses, orientations, hydrogen bond formed, and Van der Waals and charge interactions of synthesized compound 3a with enzyme transpeptidase.
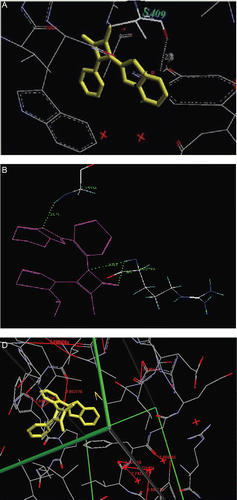
Table 2. Molecular docking showing binding score of synthesized compounds in to the active site of enzyme transpeptidase.
Table 3. Physiscal data of synthesized compounds 3a-3h.
Results and discussion
Target compounds 3a-3h ware prepared by synthetic protocol described in Scheme 1.Schiff bases (2a-2h) formation was carried out by reacting 2-(1H-benzimidazol-2-yl)benzenamine (1) with different aromatic aldehydes in the presence of sulphuric acid. Resultant Schiff bases were cyclised in to 2-azetidinones 3a-3h with chloro acetyl chloride in the presence of triethylamine in dry 1,4-dioxane. Triethylamine was used to absorb released hydrochloric acid. The signals of 1HNMR and IR were in complete agreement with the assigned structures to compounds 3a-3h. The mass spectra of these compounds displayed a molecular ion peak at appropriate m/z values which were corresponding well with the respected molecular formulas. All the compounds have given the satisfactory elemental analysis by C, H and N. All compounds 3a-3h screened, has shown good activity when compared with reference antibacterial ampicillin. Compounds had shown MIC in the range 1.0-86 μM/L. Compounds 3a, 3b, 3d and 3g were found to be potent antibacterials, with MICs 1.0-10 μM/L against Bacillus subtilis and Staphylococcus aureus. Molecular docking against enzymes transpeptidase shows binding energy (dock score) for compounds 3a, 3b, 3d, 3g and ampicillin are -11.32, -10.82, -14.83 -12.18, and 10.11 kj/moles, respectively.
Conclusion
The functionalized β-lactam compounds 3-chloro-4-substituted-1-(2-(1H-benzimidazol-2-yl)phenyl))-azetidin-2-ones 3a-3h can be easily prepared in good yields. Synthesized scaffold was screened for in vitro antibacterial activity against various strains. Solvent DMSO was incubated as control shows no antibacterial activity. All compounds screened, has shown good activity when compared with reference antibacterial ampicillin. Compounds had shown MIC in the range 1.0–86 μM/L. Compounds 3a, 3b, 3d and 3g were found to be potent antibacterials, with MICs 1.0–10 μM/L against Bacillus subtilis and Staphylococcus aureus. Docking was performed using Molecular Design Suite (MDS) v 3.5, into the 3D model of the catalytic site of enzymes transpeptidase, and to well correlate the obtained binding score with inhibitory activities of compounds. Obtained results were evaluated in terms of binding score into the catalytic site of enzymes transpeptidase, affinity and orientation of molecules in to the active site of enzyme transpeptidase. Low binding scores represent high affinity for the receptor. Compounds 3a-3h were docked in to the active site of enzyme transpeptidase. Binding energy (dock score) obtained for compounds 3a, 3b, 3d, 3g and ampicillin are -11.32, -10.82, -12.18, -14.83 and -10.11 kj/moles respectively against transpeptidase enzyme. It has found from the docking experiments that, all synthesized compounds forms hydrogen bond between oxygen of 2-azetidinone and hydroxyl group of serine 409. Further tested compounds show large charge interactions with surrounding amino acid residues in to the active site of transpeptidase. Smaller dock score (binding energy) shows more affinity towards the receptor. Compounds 3a, 3b, 3d and 3g have more affinity for receptor as well as antibacterial potential than standard ampicillin. Good correlation is found between in-silico molecular docking experiments and in vitro antibacterial activity.
Experimental
Synthesis of 2-(1H-benzimidazol-2-yl)-benzenamine (1)
In 250 ml round bottom flask, 0.01 M (1.08 g) O-phenylenediamine was taken; to it was added 20 mL of ethylene glycol. O.03 M (4.11 g) anthranilic acid and 2–3 drops of polyphosphoric acid ware added. The reaction mixture was heated on sand bath at about 190–195°C for 4 h. The reaction mixture was poured into crushed ice. Precipitate formed was filtered, washed with cold water, dried and recrystalized from ethanol. Yield: (77%). M.p: 190–192°C, Rf: -0.62 [benzene: ethyl acetate (9:1)].
General procedure for synthesis of 2-(1H-benzimidazol-2-yl)-N-[Un/substituted- benzylidene]benzenamine
Equimolar quantities of unsubstituted/substituted aromatic aldehydes and 2-(1H-benzimidazol-2-yl)-benzenamine were dissolved in 20 mL of warm dry ethanol. To it was added 1–2 drops of concentrated sulfuric acid and heated at reflux for 3 h on water bath. After standing for approximately 1 h at room temperature (r.t), the crystalline product was separated by filtration, dried.
General procedure for synthesis of 1-(2-(1H-benzimidazol-2-yl)phenyl)-3-chloro-4-[(Un)/substituted phenyl)]azetidin-2-one
To 0.01 M various 2-(1H-benzimidazol-2-yl)-N-[Un/substitutedbenzylidene]benzenamines, was added 20 ml of 1,4 dioxane. The mixture was warmed to dissolve, the resultant solution was allowed to cool, and to it was added 0.01 M triethyl amine and 0.01 M chloroacetyl chloride drop wise and with stirring. Mixture was refluxed on the boiling water bath for 4 h. Allowed to cool and filtered at pump, air dried.
1-(2-(1H-benzimidazol-2-yl)phenyl)-3-chloro-4-phenylazetidin-2-one (3a)
C22H16ClN3O, Yield: 76%, m.p: 276–278°C, IR ( KBr cm−1): 3061.06 (aromatic C-H str), 1751.42(C=O str amides),1248.67 600 (C-lCl str). 1H NMR (CDCl3): δ 4.4 (s, 1H, -NH), δ 5.4 (s, 1H, C4-H), δ 5.7 (s, 1H, C3-H), δ 6.9–8.3 (m, 13H, Ar-H). m/e: 373.
1-(2-(1H-benzimidazol-2-yl)phenyl)-3-chloro-4-(2-chlorophenyl)azetidin-2-one (3b)
C22H15Cl2N3O, Yield: 65%, m.p: 154–156°C, IR (KBr cm−i): 3232.80 (N-H stretching sec amine), 3066.06 (Aromatic C-H str ), 1749.49 (C=O str amides), 734.90 (C-Cl str). 1H NMR (CDCl3): δ 5.2 (s, 1H, C4-H), δ 5.5 (s, 1H, C3-H), δ 6.7–8.6(m, 12H, Ar-H), δ 4.2 (s,1H,-N-H) m/e:408.
In vitro anti-bacterial assay
The cultures were obtained in Mueller–Hinton Broth (Difco) for all the bacteria after 18–24 h of incubation at 37 ± 1°C. Testing was carried out in Mueller–Hinton Broth at pH 7.4 and 2-fold dilution techniques was applied. A set of tubes containing only inoculated broth was kept as controls. After incubation for 18–24 h at 37 ± 1°C, the last tube with no growth of microorganism was recorded to represent MIC expressed in μM/L. Ampicillin was used as standard drug.
In-silico molecular docking
Molecular docking of compounds 3a–3h in to the three-dimensional X-ray structure of transpeptidase was carried out using the molecular design Suite (MDS) software package (v. 3.5). The protein–ligand complex was constructed based on the X-ray structure transpeptidase. All compounds were built using Chem Draw Ultra v. 8.0 and minimized using the Merck Molecular Force Field. Keeping program parameters to their default values, the docking was performed using Molecular Design Suite (MDS) into the 3D model of the catalytic site of transpeptidase. Genetic algorithm implemented in MDS has been successfully employed to dock inhibitors into the catalytic site of the enzyme. The obtained binding score was found to be in good agreement with inhibitory activities of respective compounds. Comparative docking experiments of compounds 3a–3h with known antibacterial agent such as ampicillin were performed. Obtained results were evaluated in terms of binding score in to the catalytic site enzyme transpeptidase.
Acknowledgements
We acknowledge our thanks to SAIF Chandigarh, Punjab for spectral analysis. We are grateful to SAIF Punjab University, Chandigarh for the help in undertaking NMR and Mass spectra.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Holten KB, Onusko EM. 2000;62:611.
- Chambers HF. Penicillin-binding protein-mediated resistance in pneumococci and staphylococci. J Infect Dis 1999;179 Suppl 2:S353–S359.
- Hart CA. “Antibiotic resistance: an increasing problem?” BMJ. 1998;316(7140) 1255–1256 Kozo, A.; Kazuhiro, Masayuki, K.; U.S. 6,815,455, 2001; Chem Abstr 2001;134:86247.
- Jarrahpour A, Shekarriz M. Asymmetric Synthesis and Antimicrobial Activity of Some New Mono and Bicyclic β-Lactams, Molecules, 2004;9:939.
- Rossi S. Australian Medicines Handbook. (2004). Adelaide: Australian Medicines Handbook.
- Chhajed SS, Upasani CD, Jagdale SB. “Synthesis, Physicochemical Properties and Antimicrobial Activity of Some 2-Substituted analogues of Benzimidazoles, J Pharmacy Res, 2010;3:1250–1253.
- Spratt BG. Properties of the penicillin binding proteins of Escherichia coli K12. J Biochem, 1977;14:342–352
- Desai KG, Desai KR. Green route for the heterocyclization of 2-mercaptobenzimidazole into beta-lactum segment derivatives containing -CONH- bridge with benzimidazole: Screening in vitro antimicrobial activity with various microorganisms. Bioorg Med Chem 2006;14:8271–8279.
- Chhajed SS, Bastikar VA, Haldar A, Ingle VN, Upasani CD, Wazalwar SS, Synthesis and molecular modeling studies of 3-chloro-4-substituted-1-(8-hydroxy-quinolin-5-yl)-azetidin-2-ones as novel anti-filarial agents Bioorg. & Med. Chem. Let, 2010;20:3640–3644.
- Chhajed SS, Hiwanj P, Bastikar V, Upasani C, Dhake A, Mahajan, Structure Based Design and In-Silico Molecular Docking Analysis of Some Novel Benzimidazoles J Chem Tech Res, 2010;2:1135.
- Chhajed SS, Upasani CD, Bastikar VA, Mahajan NP, Synthesis, Physicochemical Properties and Biological Evaluation Of Some Novel 5-[2-methyl/(un) Substituted Phenylethylideneamino] Quinolin-8-ols, J Pharmacy Res, 2010;3:1192.
- Chhajed SS, Padwal M, Antimicrobial Evaluation of Some novel Schiff and Mannich bases of Isatin and its derivatives with quinolin Int J Chem Tech Res, 2010;2:209.
- Jiaxi X, Issue 5th Eurasian Conference on Heterocyclic Chemistry ARKIVOC, 2009;21:22–23.
- Kelly J, Kuzin A. A point mutation leads to altered product specificity in β-lactamase catalysis J Mol Biol 1995;254:223.
- Kelly JA, Kuzin AP. The refined crystallographic structure of a DD-peptidase penicillin-target enzyme at 1.6 A resolution. J Mol Biol 1995;254:223–236.