Abstract
Src family kinase (SFK) is a family of protein tyrosine kinases that play important roles in the development of various cancers. Here, we showed that a naturally occurring inhibitory factor of SFK can be extracted from the rat brain. This inhibitor strongly suppressed the activity of SFKs including Lck and Fyn. It did not inhibit other protein tyrosine kinases including Wee1 or serine/threonine kinases Mst2, Cdk5/p25, Cdk5/p35, and Cdk2/cyclin A. The inhibitor was not an ATPase, a phosphatase that dephosphorylates substrates of the SFK reaction, or a protease that degrades SFKs. Activity of mutant Lck with C-terminal tyrosine substituted with phenylalanine was also suppressed by the inhibitor to a similar extent of wild-type Lck, indicating that the inhibitor was not CSK. Gel filtration chromatography indicated that the molecular size of the prevalent form of this inhibitor was approximately 44 kDa.
Keywords::
Introduction
Src family kinase (SFK) is a family of nonreceptor protein tyrosine kinases (e.g., Src, Fyn, Lck). Src family kinase activity was reported to be elevated in various human cancers, including breast cancer and colon cancers (reviewed in Ref. Citation1). This increased Src activity has been shown to play important roles in the development of human cancers. Src exerts its effect through multiple signalling pathways including Shc/Grb2/ERK2 pathway, STATs (Signal Transducer and Activator of Transcription) pathway, and focal adhesion kinase (FAK) pathwaysCitation2.
Src activity is negatively regulated by CSK, a ubiquitously expressed protein kinaseCitation3, via phosphorylation at Y530 of Src. In addition, CSK homologous kinase (CHK) has been shown to perform a similar role in brain and hematopoietic cellsCitation4. Both CSK and CHK have been shown to be capable of phosphorylating Lck and repressing its kinase activity. Phosphorylated Y530 interacts with the SH2 domain of Src, resulting in the suppression of its kinase activityCitation5. However, CSK family kinase has also been reported to be elevated in cancer cells, where Src was activatedCitation6,Citation7, suggesting that there might be potential negative regulators yet undiscovered for Src kinase activity.
In this article, using tissue extraction, DEAE Sepharose Fast Flow chromatography, dialysis and gel filtration chromatography, we found a 44 kDa component in rat brain, which specifically inhibits Src family kinases. The nature of this inhibitor has been explored and described in this article.
Materials and methods
Radioisotope and antibodies
[γ-32P]ATP (Adenosine-5′-triphosphate) and HRP-conjugated (horseradish peroxidase) secondary antibodies were from Amersham. Anti-Fyn and anti-Lck polyclonal antibodies raised against peptides corresponding to amino acids at the carboxyl terminal of human Fyn and Lck, respectively, were made in our laboratory.
Proteins and enzymes
His-Mst2, His-Cyclin A, GST-Lck, GST-Fyn, GST-Cdk2, GST-Cdk5, GST-p25, GST-p35, GST-Wee1 were prepared in our laboratory. Molecular weight markers were from Bio-Rad.
Molecular biology reagents
GST-fusion expression vector and Escherichia coli MM294 and BL21 were from Amersham. Restriction endonucleases, restriction buffers, DNA ladder were from Gibco-BRL Life Technologies. Gel extraction kit was from QIAGEN. Oligonucleotides were synthesized by Gibco-BRL Life Technologies. All the peptides were synthesized by Peptide Facility, the University of Calgary, Canada.
Other chemicals and materials
Antipain, leupeptin, aprotinin, benzamidine, and glutathione were from Sigma. Phenylmethylsulphonyl fluoride (PMSF), dithiotreitol (DTT), isopropyl β-d-thiogalactoside (IPTG), and soybean trypsin inhibitor were from Gibco-BRL Life Technologies. Enhanced Chemiluminescence (ECL) Western Blotting Detection Reagents, acrylamide, glutathione Sepharose 4B, DEAE Sepharose Fast Flow, FPLC Superose 6 were from Amersham. Bio-Rad Protein Assay Reagent was from QIAGEN. Immun-Blot PVDF membrane was from Bio-Rad. P81 Phosphocellulose paper was from Whatman. X-ray films were from Fuji.
Construction of mutant Lck
Site-directed mutagenesis at the C-terminus of Lck was performed using QuikChange Site-Directed Mutagenesis Kit (Stratagene). The mutation primers 5′-CACTTAATGGGAAACTTGGCCC-3′ and 5′-CCTGACCACTGGCCAAGAAGGC-3′ have been subjected to FPLC purification. The Tm of polymerase chain reaction (PCR) is 80°C. The amount of dsDNA of rat pGEX4T-1-Lck [rat and human Lck protein sequences are highly homologous (99% homology)] in the PCRs is 50 ng. After one cycle at 95°C for 30 s, 12 cycles at 95°C for 30 s, 55°C for 1 min, and 68°C for 13 min are conducted. Digesting of DNA products, transforming into Epicurian Coli XL1-Blue supercompetent cells, and selection of the positive clones were all conducted as described in the instruction manual. The positive clone of mutant Lck DNA was checked for DNA insert size with agarose gel electrophoresis and then sequenced to confirm that mutation was successful.
Transformation and purification of plasmid
Transformation was performed by Transformation and Storage Solution (TSS) methodCitation8. The DNA plasmids were purified by Wizard Plus Minipreps DNA Purification System Kit (Promega) according to the manufacturer’s protocol.
Expression and purification of GST-fusion proteins
The recombinant Fyn/pGEX-2T and Lck/PGEX-4T-1 constructs were expressed in E. coli MM294 and BL-21 (DE3) strain, respectively. GST-fusion proteins were purified as describedCitation9.
Determination of protein concentration
Protein concentration was determined by Bradford methodCitation10.
SDS-PAGE and immunoblotting
SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) and immunoblotting were performed as describedCitation11.
Assay of kinase activity
Lck and Fyn
Lck or Fyn was incubated in 30 μl mixture containing 20 mM MOPS (3-[N-Morpholino]propane-sulfonic acid), pH 7.5, 10 mM MgCl2, 0.1 mM Na3VO4, 0.5 mM cdc2(6-20) peptide, and 100 μM [γ-32P]ATP at 30°C for 20 min. The reaction was terminated by adding 7.5 μl of 50% trichloroacetic acid (TCA) and 12.5 μl of 0.1 mg/ml bovine serum albumin (BSA), followed by centrifugation at 12,000 × g for 5 min. Thirty microliters of the reaction was spotted to P81 paper squares and the amount of phosphorylation was counted by a Scintillation counter. Lck and Fyn kinases used were quite stable (no apparent change in the kinase activities after incubation at 30°C for up to 40 min was noted).
Mst2, Cdk2 and Cdk5, and Wee1
Assays were performed as above except using different substrates: 100 μM cdc2(6-20) peptide (Mst2); 100 μM histone H1(9-18) peptide (Cdk2 and Cdk5). For Wee1, the substrate is 0.5 mM cdc2(4-24), and the reaction was terminated by adding 10 μl of 100% acetic acid. For assay of Cdk2 and Cdk5, a reconstitution with their cyclin activators was performed first to achieve stable kinase activity before the kinase assay: GST-Cdk2, His-cyclin A, and bovine thymus-purified CAK were reconstituted in the reconstitution buffer containing 30 mM MOPS, pH 7.4, 10 mM MgCl2, 150 mM NaCl, 0.5 mM ATP, and 1 mg/ml bovine serum albumin for 1 h at 30°C; GST-Cdk5 and GST-p25 were reconstituted in phosphate-buffered saline (PBS) buffer containing 1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM DTT, and 1 mg/ml bovine serum albumin for 1 h at 30°C. The Cdk5/p35 was lysate of insect cell (lysis buffer: see Protein fractionation; 150,000 × g supernatant) co-transfected by cdk5/p35, incubated in the reconstitution buffer containing 1 mg/ml BSA at 30°C for 10 min.
Kinase inhibition assay
Protein kinase was added with column fractions or other samples, in the 25 mM HEPES (4-(2-hydroxyethyl)-1-piperazine-ethanesulfonic acid) buffer (pH 7.2) for 20 min, unless otherwise stated. Then, kinase activity was assayed.
Phosphatase test
Kinase was added in 30 mM MOPS, pH 7.5, 10 mM MgCl2, 0.1 mM Na3VO4, 0.5 mM cdc2(6-20) peptide, and 100 μM [γ-32P]ATP at 30°C for 20 min for phosphorylation of substrate. Then, tubes were heated at 95°C for 7 min and cooled on ice for 10 min. Column fraction or other samples were added and incubated at 30°C for 20 min. The reaction was stopped by addition of 50% (v/v) TCA. Aliquots of the supernatant were spotted on P81 paper squares. The papers were washed with 0.3% phosphoric acid, and the incorporation of the 32P-phosphate into the histone peptide was measured by liquid scintillation counter.
ATPase test
The sample was preincubated with 100 μM [γ-32P]ATP (300 cpm/pmol) and 100 μM cdc2(6-20) peptide in 30 mM MOPS, pH 7.4, 10 mM MgCl2, at 30°C for 20 min. The sample was heated at 95°C for 2 min, cooled on ice for 5 min, and then added with kinase to incubate at 30°C for 6 min. The reaction was stopped by addition of 50% (v/v) TCA. P81 paper spotted with aliquots of the supernatant was washed extensively with 0.3% phosphoric acid before the incorporation of the 32P-phosphate into the cdc2(6-20) measured by liquid scintillation counter.
Protein fractionation
Tissue extract
All procedures were carried out at 4°C. Ten grams of brain tissue from Sprague-Dawley strain rats were lysed in 20 ml lysis buffer (25 mM HEPES, pH 7.2, 1 mM EDTA, 1 mM DTT, 0.6 mM PMSF, 2 μg/ml antipain, 2 μg/ml leupeptin, 1 μg/ml aprotinin, 0.3 mg/ml benzamidine, and 0.1 mg/ml soybean trypsin inhibitor) by pestle tissue grinder. The crude homogenate was centrifuged at 12,000 × g for 30 min, and the supernatant was further centrifuged at 120,000 × g for 45 min to obtain the clear tissue extract (supernatant of 120,000 × g). The study was approved by Animal Care and Use Committee.
DEAE column chromatography
Tem milliliters of the clear tissue extract was loaded at 1 ml/min onto a 3-ml DEAE Sepharose Fast Flow column pre-equilibrated with buffer A (25 mM HEPES, 1 mM DTT, 1 mM EDTA, 1 mM PMSF, 1 μg/ml each of antipain, leupeptin and aprotinin, 2 μg/ml trypsin inhibitor, and 0.3 mg/ml benzamidine). Then, the column was washed with buffer A at 1 ml/min until the UV trace of the effluent dropped to the baseline steadily. The column was eluted with a linear salt gradient (0–0.5 M NaCl) in buffer A at 0.7 ml/min in 50 min. A total of 0.5 ml per fraction was collected.
FPLC Superose 6 Gel Filtration Column Chromatography
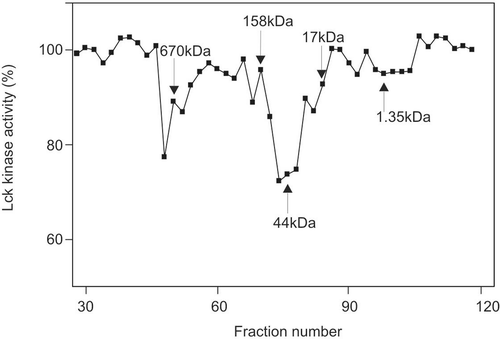
Pooled samples from a DEAE Sepharose Fast Flow column were dialyzed and centrifuged at 10,000 × g for 20 min. Two hundred microliters of the supernatants or molecular weight standards were then applied to an FPLC Superose 6 gel filtration column equilibrated with the lysis buffer containing 0.15 M NaCl and 0.2 mg/ml BSA. The column was eluted at 0.4 ml/min and 0.3 ml per fraction was collected. The molecular standards were thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa), and vitamin B-12 (1.35 kDa).
Results and discussion
Profile of inhibition of GST-Lck by DEAE column fractions
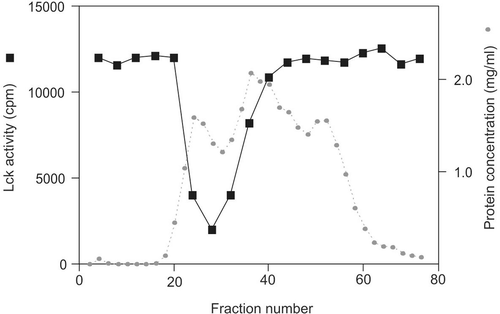
Clear tissue extracts from the rat brain were applied to a 3-ml DEAE Sepharose Fast Flow column. After thorough washing, the column was eluted with a linear gradient of 0–0.5 M NaCl in the homogenization buffer. Two microliters of each fraction were assayed for its activity to downregulate Lck kinase activity in a kinase inhibition assay, which was performed with a 20-min pre-incubation of the fractions with Lck kinase, followed by another 20 min in the presence of ATP and peptide substrate cdc2(6-20) for assay of kinase activity (). The inhibitory activity against GST-Lck came out with its peak at NaCl concentration of 0.065 M. The loading was approximately 100 mg, and the protein concentration at the peak was 1.3 mg/ml. A strong potency of inhibition was displayed in the fractions 27–29: 2 μl of fraction (~2.6 μg of total protein) produced 85% inhibition of Lck activity.
Figure 1. GST-Lck inactivation profile of DEAE Sepharose Fast Flow column eluted proteins. Two microliters of fraction sample was incubated with GST-Lck for 20 min followed by 20 min of kinase assay against cdc2(6-20) peptide substrate. This is a representative profile of three reproducible experiments.
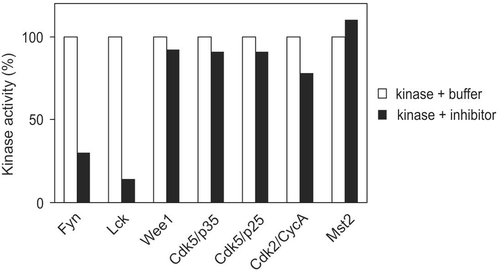
Inhibitory specificity
Peak fractions of GST-Lck inactivation (fraction 27–29) from the DEAE Sepharose Fast Flow column were pooled and dialyzed (MW cut off: 8 kDa) against homogenization buffer. While 1.3 μl (~1.6 μg protein) of before-dialysis sample caused 50% inhibition on the Lck kinase activity, 3.6 μl (4.3 μg protein) of the dialyzed sample exhibited the same effect, indicating that approximately 40% of the inhibitory activity was retained after the dialysis. The remaining inhibitory factor after dialysis should be larger than 8 kDa in molecular weight. It is possible that degradation of the inhibitory factor accounted for some of the inhibitory activity lost in the dialysis.
To investigate the specificity of this inhibitory factor, several protein kinases were employed. These kinases include His-Mst2, GST-Wee1 and GST-Cdk2/His-cyclin A, GST-Cdk5/GST-p25, His-Cdk5/His-p35, GST-Fyn, and GST-Lck (). While the dialyzed inhibitor sample reduced 86% of GST-Lck activity, it only displayed marginal effect on activities of Cdk5/p25 (9%), Cdk5/p35 (9%), and Wee1 (8%). Its impact on Cdk2/cyclin A was also much less effective as compared with Lck. Furthermore, GST-Fyn was also a very effective target for this inhibitor, with an inhibition of 70%.
Characterization of the inhibitory activity
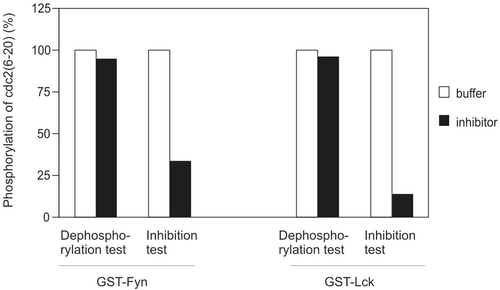
A phosphatase could dephosphorylate phosphorylated substrate of the Src kinase reaction and lead to a decline in the radioactive labelling (cpm) of the substrate in the assay. To examine this possibility, the target protein kinase (GST-Fyn or GST-Lck) was first incubated with [γ-32P]ATP/Mg2+ and peptide substrate for 20 min to allow the substrate to be phosphorylated, and the activity of Src family kinase in the reaction was then destroyed by heat treatment (95°C for 7 min). After the tubes had been cooled down, the inhibitory sample was added to the reaction to incubate at 30°C for 20 min, followed by counting the γ-32P incorporation in the phosphopeptide. While the sample could reduce the activity of GST-Fyn by 66.5% in the inhibition test, it had little effect on dephosphorylating the cdc2(6-20) peptide phosphorylated by GST-Fyn (5.0%) (). Similarly, while GST-Lck kinase activity was reduced by 86.2% after inhibition, phosphatase presence in the sample could only account for less than 4% of the reduction in radioactive labelling. These data indicate that the identity of the inhibitory activity was not a phosphatase that dephosphorylates phosphorylated peptide substrates. In another set of experiment, we also employed various phosphatase inhibitors in the kinase inhibition assay including sodium orthovanadate (Na3VO4, 50 μM), okadaic acid (40 μM), fenvalerate (8 μM), but they did not block the kinase inactivation (data not shown), which further excluded the existence of a phosphatase accounting for the observed kinase inhibition.
Figure 3. Possibility of phosphatase. Buffer or inhibitor sample was assayed for their ability to dephosphorylate phosphor-cdc2(6-20) or inhibit kinase activity of GST-Fyn and GST-Lck. The amount of kinase and inhibitor were identical in phosphatase test and kinase inhibition test. Both the time for substrate dephosphorylation (phosphatase test) and preincubation (kinase inhibition test) were 20 min. This result is representative of three reproducible experiments.
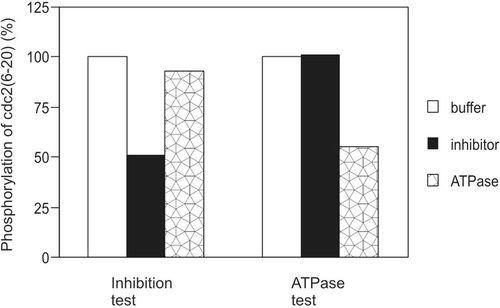
To identify whether the inhibitory activity was an ATPase that hydrolyses ATP in the Src kinase assay, we preincubated the inhibitory sample with ATP/Mg2+ for 20 min, the same amount of time of kinase/sample preincubation that produced the inhibition in the corresponding inhibition test. Then, the tubes were heated at 95°C for 2 min to destroy the inhibitory activity. Following cooling the sample on ice for 5 min, the kinase was added to initiate the kinase reaction at 30°C for 6 min for the phosphorylation of the peptide substrate. In a kinase inhibition assay, an inhibitory sample reduced peptide substrate phosphorylation by GST-Lck by 49%, while an ATPase sample by only 7% (). On the other hand, while the ATPase sample destroyed ATP by 45%, the inhibitory sample did not destroy ATP at all (ATPase test, ). These data demonstrated that the inhibitory activity was not an ATPase.
Figure 4. Possibility of ATPase. Buffer, inhibitor, or positive ATPase sample was assayed for their ability to inhibit substrate phosphorylation by GST-Lck or to hydrolyse ATP. The amount of kinase, inhibitor, and positive ATPase were identical in the kinase inhibition test and the ATPase test. The time of preincubation for both the kinase inhibition test and ATPase test were 20 min. This result is representative of three reproducible experiments.
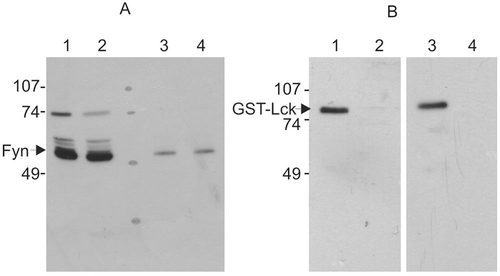
Proteases may degrade and inactivate Src family kinases. However, the homogenization buffer, elution buffer, and dialysis buffer we used all contained a wide range of protease inhibitors, which suggest that the apparent inhibitory activity on Src kinase reaction was not due to a protease. To further verify this, a Western blot analysis of the treated kinases was examined (). Incubation of the inhibitory factor with Fyn (purified from bovine thymus) reduced the activity of Fyn by 70%, but the amount of the Fyn was not changed (). Similarly, incubation of the inhibitory factor with GST-Lck led to a decrease of approximately 12-fold in the kinase activity; however, there was no change in amount of the 82 kDa full-length GST-Lck ().
Figure 5. Western blot of protein kinases treated with the inhibitory sample. Protein kinases were incubated with lysis buffer or inhibitory sample for 20 min and then incubated for 20 min in the presence of ATP and cdc2(6-20) peptide substrate. Reaction was terminated by heat denaturation. (A) Purified bovine Fyn with buffer A (lanes 1 and 3) and inhibitor sample (lanes 2 and 4). The amount of Fyn protein sample loaded was 500 ng (lanes 1 and 2) and 50 ng (lanes 3 and 4). The Fyn kinase activity after incubation with inhibitor sample was 30% of that treated with buffer A. (B) GST-Lck with buffer A (lanes 1 and 2) and inhibitor sample (lanes 3 and 4). The amount of GST-Lck protein loaded on each track was 40 ng (lanes 1 and 3) and 13 ng (lanes 2 and 4). The GST-Lck kinase activity treated with inhibitor sample was 8% of that treated with buffer A. This result is representative of three reproducible experiments.
CSK is a 50 kDa protein kinase that inactivates Src family protein kinases by phosphorylating their C-terminal tyrosine (Tyr531 in Fyn, Tyr505 in Lck). Comparison of the activities of highly purified phosphorylated and dephosphorylated Src shows that phosphorylation of the C-terminal tyrosine inhibits activity by greater than 98% and overrides any stimulatory effects due to phosphorylation of Tyr416 in the catalytic domainCitation12.
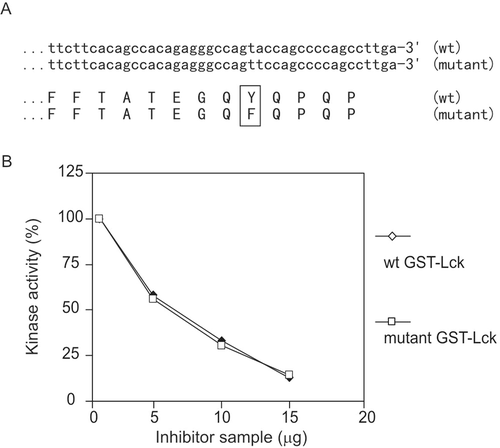
To examine whether the inhibitory factor was CSK, a mutant Lck was constructed with the C-terminal tyrosine mutated to phenylalanine (). The dose-dependent curve shows that the activities of both the mutant GST-Lck and wild-type GST-Lck were suppressed by the inhibitory factor, and with almost identical efficiency (). This indicates that the C-terminal tyrosine of Lck was not crucial in the inhibition; therefore, CSK was not the cause of the inhibition. For the same reason, CHK was not the cause of the inhibition either.
Figure 6. CSK test. (A) Alignment of nucleotide sequences at 3′ of wild-type and mutant Lck and corresponding amino acid sequences at C-terminus. The full-length sequences were obtained and identical except at the 3′ (C-terminus), which is shown. (B) Dose-dependence comparison of inhibition on wild-type and mutant GST-Lck. This result is representative of three reproducible experiments.
Determination of molecular size of the inhibitory activity
To determine the molecular size of this inhibitor, inhibitory sample from DEAE column was loaded on a gel filtration column Superose 6, along with a molecular standard. Two main peaks of inhibitory activity were displayed, with the prevalent activity approximately 44 kDa, and the minor activity over 670 kDa ().
Effects of preincubation and salt on Lck inhibition
The routine assay of the Lck kinase inhibition includes a 20-min preincubation of Lck with the inhibitor sample, followed by kinase assay in the presence of ATP and peptide substrate cdc2(6-20). We also tested the reduced preincubation time of 10 and 5 min, while keeping the kinase assay unchanged. While a 94.2% inhibition of Lck activity was achieved at 20 min preincubation, less inhibitions (78.5% and 62.7%) were achieved at 10 and 5 min, respectively.
We also tested whether high concentration of salt could affect the inhibition of Lck by the inhibitor. While an Lck inhibition of 62.4% in a routine inactivation assay was achieved, the inhibition was decreased to 38.0% when 0.5 M NaCl was added at preincubation. The reduction of Lck inhibition with 0.5 M NaCl suggests that high concentration of salt could disrupt the interaction between Lck and the inhibitor.
In summary, this report has demonstrated the existence of a novel negative regulator of Src family kinase in the brain tissue of rats. Src family kinase is abundant in brain tissues and plays important roles in neuronal signalling. The existence of this inhibitor in the brain is compatible to the necessity of tight regulation of Src family kinase in the brain. Lck is most abundantly expressed in cells of lymphoid lineage, especially T lymphocytesCitation13. To further investigate the relationship between Lck and the inhibitor, we have also extracted rat thymus and bovine spleen tissues and have obtained inhibitor with similar properties. This suggests that the expression of this inhibitor might be universal. More importantly, since Src family kinase plays an essential role in the development of various cancers, novel natural Src inhibitors may be potentially critical in the therapeutic intervention in cancers. It is our ultimate goal to fully identify this inhibitor and explore the detailed mechanism involved in the regulation of Src family kinases and the functional roles it play in development of cancers.
Declaration of interest
The authors report no conflicts of interest.
References
- Bjorge JD, Jakymiw A, Fujita DJ. Selected glimpses into the activation and function of Src kinase. Oncogene 2000;19:5620–5635.
- Sayeski PP, Ali MS. The critical role of c-Src and the Shc/Grb2/ERK2 signaling pathway in angiotensin II-dependent VSMC proliferation. Exp Cell Res 2003;287:339–349.
- Okada M, Nakagawa H. A protein tyrosine kinase involved in regulation of pp60c-src function. J Biol Chem 1989;264:20886–20893.
- Avraham S, Jiang S, Ota S, Fu Y, Deng B, Dowler LL et al. Structural and functional studies of the intracellular tyrosine kinase MATK gene and its translated product. J Biol Chem 1995;270:1833–1842.
- Liu X, Brodeur SR, Gish G, Songyang Z, Cantley LC, Laudano AP et al. Regulation of c-Src tyrosine kinase activity by the Src SH2 domain. Oncogene 1993;8:1119–1126.
- Bénistant C, Bourgaux JF, Chapuis H, Mottet N, Roche S, Bali JP. The COOH-terminal Src kinase Csk is a tumor antigen in human carcinoma. Cancer Res 2001;61:1415–1420.
- Watanabe N, Matsuda S, Kuramochi S, Tsuzuku J, Yamamoto T, Endo K. Expression of C-terminal src kinase in human colorectal cancer cell lines. Jpn J Clin Oncol 1995;25:5–9.
- Chung CT, Miller RH. Preparation and storage of competent Escherichia coli cells. Meth Enzymol 1993;218:621–627.
- Tang D, Chun AC, Zhang M, Wang JH. Cyclin-dependent kinase 5 (Cdk5) activation domain of neuronal Cdk5 activator. Evidence of the existence of cyclin fold in neuronal Cdk5a activator. J Biol Chem 1997;272:12318–12327.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254.
- Zhu S, Bjorge JD, Fujita DJ. PTP1B contributes to the oncogenic properties of colon cancer cells through Src activation. Cancer Res 2007;67:10129–10137.
- Stover DR, Liebetanz J, Lydon NB. Cdc2-mediated modulation of pp60c-src activity. J Biol Chem 1994;269:26885–26889.
- Marth JD, Peet R, Krebs EG, Perlmutter RM. A lymphocyte-specific protein-tyrosine kinase gene is rearranged and overexpressed in the murine T cell lymphoma LSTRA. Cell 1985;43:393–404.