Abstract
Ranitidine is an antagonist of histamine-2 (H2) receptor. It is employed to treat peptic ulcer and other conditions in which gastric acidity must be reduced. Sucrase is a hydrolytic enzyme that catalyzes the breakdown of sucrose to its monomer content. A liquid of yeast sucrase was developed for treatment of congenital sucrase-isomaltase deficiency (CSID) in human. In this study, the effect of ranitidine on yeast sucrase activity was investigated. Our results showed that ranitidine binds to sucrase and inhibits the enzyme in a noncompetitive manner. The Ki and IC50 values were measured to be about 2.3 and 2.2 mM, respectively. Fluorescence measurement showed conformational changes after binding of ranitidine to the enzyme. The fluorescence spectra showed that ranitidine could bind to both free enzyme and enzyme–substrate complex, which was accompanied with reduction of emission intensity and red shift production.
Keywords::
Introduction
Histamine-2 (H2) receptor blockers, such as ranitidine and cimetidine, are a group of drugs that work by reducing the acid secretion in stomachCitation1. Ranitidine and cimetidine (under the trade names Zantac and Tagamet, respectively) are used to treat ulcers in gastrointestinal tractCitation2. Most of the drugs have shown to induce side effects in body. Ranitidine reduces the activity of some enzymes residing in renal brush border, such as hydrolytic enzymesCitation3, and also duodenal mucosal enzymes in patients with peptic ulcerCitation4. Sucrase is a hydrolytic enzyme that breaks down sucrose into glucose and fructose. The human gastrointestinal system has a kind of sucrase known as sucrase-isomaltase that resides in the apical surface of intestinal cells. Yeast sucrase has been shown to be used for patients with congenital sucrase-isomaltase deficiency (CSIDCitation5,Citation6). Some drugs can inhibit the sucrase activity. Nojirimycin, deoxynojirimycin, and acarbose have shown competitive inhibition of intestinal sucraseCitation7. Castanospermine, a drug that is mainly used for inhibition of α-glucosidase, induces noncompetitive inhibition on sucrase activityCitation8. Previous report also indicated that codeine binds to yeast sucrase and inhibits the enzyme via noncompetitive inhibitionCitation9. In this study, the effect of drug ranitidine on the activity of sucrase has been studied and kinetics of binding and structural changes were reported. The use of sacrosidase for treatment of CSID induces some side effects in gastrointestinal tract, which are similar to symptoms caused by CSID (such as stomach ache, vomiting, diarrhea, nausea, and constipation). As ranitidine is used to treat gastrointestinal disorders, this report may guide physicians to administer ranitidine in patients under sacrosidase use, with precaution.
Materials and methods
Yeast sucrase (3.2.1.26) was obtained from Fluka Company. Ranitidine and cimetidine were purchased from Sigma Company. All the chemicals used for the enzyme assay and buffer preparation were of reagent grade and obtained from Merck Company.
Enzyme activity
Enzyme activity was measured according to the method reported in Ref. 9. Sucrase solution (0.5 mg/ml) was prepared by dissolving the enzyme in distilled water. The optimum pH for enzyme activity was determined to be 4.5; however, the enzyme is more stable in pH 7Citation10. As a matter of fact, ranitidine has been reported to be unstable in acidic or basic solutionsCitation11; therefore, the enzyme assay was performed in 0.1 M phosphate buffer at pH 7.
The reaction was started by adding 100 μl of sucrase solution to the test tubes containing different concentrations of sucrose (2.5–75 mM). For the assays done in the presence of ranitidine, appropriate amounts of ranitidine were added to the test tubes from the stock solution (50 mM). The final volume in each test tube was always 2 ml and the concentration of ranitidine varied from 0.5 to 3 mM.
Sucrose hydrolyzing activity of sucrase was determined by measuring the rate of liberation of glucose and fructose from sucrose and measured by the ferricyanide method for determination of reducing sugars.
The enzyme was incubated with sucrose for 1, 3, 5, 10, and 15 min in the presence and the absence of ranitidine. The activity of enzyme was stopped at the mentioned times by incubating the solution at 80°C. The ferricyanide colorimetric method was performed for the enzyme activityCitation12 using Perkin-Elmer visible spectrophotometer.
The results were the average of at least three separate experiments and were expressed as mean ± standard deviation.
Fluorescence measurements
The fluorescence spectra were taken on a Perkin-Elmer SL45 fluorometer. The excitation wavelength was 285 nm and the emission spectra were recorded in a range between 310 and 400 nm. The enzyme solution was prepared with a concentration of 0.04 mg/ml in 0.1 M phosphate buffer (pH 7). Sucrose was used with a concentration of 30 mM, and ranitidine concentration was 6.5 μM.
Results
Inhibition of sucrase activity by ranitidine
The effect of ranitidine and cimetidine on sucrose activity was monitored. Although both these drugs have nearly the similar structure and effect, no change in the enzyme activity was observed after using different concentrations of cimetidine. In contrast, ranitidine affected the sucrase activity significantly. So our measurements were focused on ranitidine and its effects on sucrase activity.
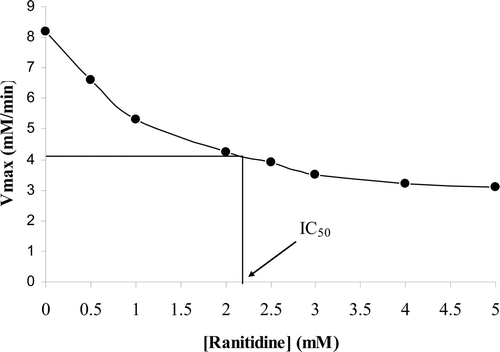
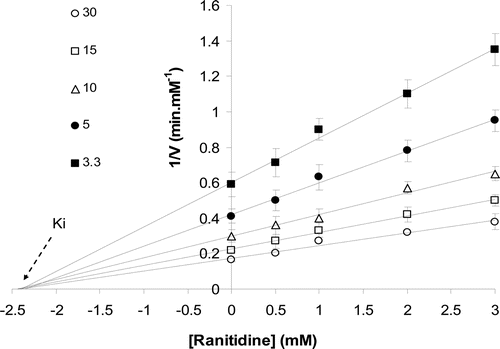
The reaction rate of enzyme on sucrose (substrate) was determined in the absence and the presence of ranitidine by detecting the formation of products (glucose and fructose) at 410 nm. The incubation of enzyme with sucrose continued to pass into the steady state and continued to reach the plateau (substrate depletion). The Michaelis–Menten analysis was applied to determine the reaction rate of the pure enzyme (). Since sucrase is more stable in pH 7, further kinetics experiments were done at pH 7. Edie–Hofstee plot was employed to determine the kind of inhibition in the presence of different concentrations of ranitidine (). A noncompetitive inhibition in sucrase was observed after different concentrations of ranitidine were added to the enzyme and substrate solution.
Figure 1. Michaelis–Menten plot shows that above 30 mM sucrose concentration, the enzyme reaches to zero-order reaction.
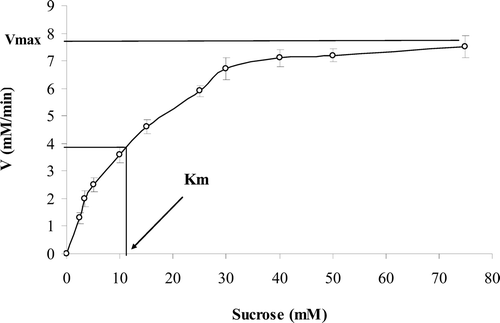
Figure 2. The pattern of Eadie–Hofstee plot showed noncompetitive inhibition after binding of ranitidine to the enzyme. The slope of the lines is the same and determines a constant Km, but intercepts on the ordinate were different, which indicate that the Vmax of the enzyme was not constant in the presence of different concentration of ranitidine (0–3 mM).
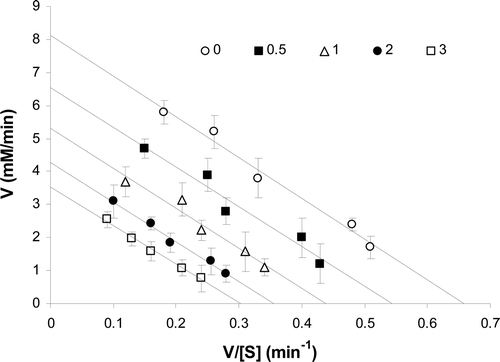
The Km (11.5 mM) of enzyme was constant in the presence of various concentrations of ranitidine (), while its Vmax decreased by increasing ranitidine concentration (). The concentration of ranitidine producing 50% inhibition of maximal activity (IC50) was determined to be about 2.2 mM (). According to Dixon plot, the inhibition constant (Ki) of ranitidine was obtained to be 2.3 mM ().
Structural changes
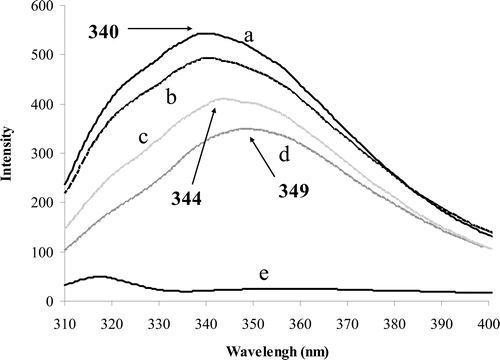
To ensure the purity of the enzyme, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed which showed only a single protein bond. Fluorescence measurements were done for determination of conformational changes occurred after binding of substrate and inhibitor to the enzyme. The excitation was performed at 285 nm for exciting tryptophan residues and the emission was recorded at a region between 310 and 400 nm. Excitation of pure enzyme showed a peak at 340 nm (, graph A). Binding of substrate to the enzyme reduced the peak intensity without any shift (, graph B). Addition of inhibitor to pure enzyme decreased the intensity of peak, which was accompanied by a red shift to 344 nm (, graph C). Addition of inhibitor to enzyme–substrate complex also decreased the peak intensity and induced a red shift to 344 nm (, graph D).
Figure 5. Fluorescence spectra obtained from excitation at 285 nm and emission range of 310–400 nm. The pure enzyme (0.04 g/ml) induces a peak at 340 nm (a). Binding of substrate (30 mM) to the enzyme reduced the peak emission intensity (b). Binding of ranitidine (6.5 μM) to free enzyme also decreased the emission intensity and induced a red shift to 344 nm (c). Binding of ranitidine to enzyme–substrate complex also induced red shift (d). Pure ranitidine (e).
Discussion
The yeast sucrase has been proposed to be applied for treatment of patients with CSIDCitation6. Sacrosidase (a liquid produced from Saccharomyces cerevisiae) is used in these patients to lessen the outcomes of sucrose deficiency 95). Several drugs induce counter effects when administered simultaneously with sacrosidase. In this case, some drugs such as nojirimycin, deoxynojirimycin, and acarbose have shown competitive inhibition of intestinal sucraseCitation7.
In the present study, the effect of ranitidine on sucrase activity was investigated. Since the stability of the enzyme and inhibitors is achieved at pH 7, most of the enzyme kinetics and structural studies were performed at this pH.
There are few reports regarding the effect of therapeutic drugs on sucrase activityCitation13. Codeine and scopolamine are the common drugs that have been reported to inhibit the yeast sucrase activityCitation9,Citation14. Some drugs such as gentamycin increase rat intestinal sucrase activityCitation15, whereas nonsteroidal anti-inflammatory drugs (NSAIDs) decrease intestinal sucrase activityCitation16. A recent report has been shown the crystal structure of the N-terminal catalytic domain of sucrase–isomaltase enzyme and has determined that some chemicals, such as kotalanol, inhibited the enzyme by binding to this domainCitation17.
Our results showed that ranitidine binds to sucrase and inhibit its activity by noncompetitive inhibition. Increasing ranitidine concentration resulted in a decrease in Vmax, while Km of the enzyme remained constant. The measurement of Ki and IC50 values of the drug showed that ranitidine binds with moderate affinity to the enzyme. On the other hand, cimetidine, the other H2 blocker drug, had no inhibitory effect on sucrase. Some drugs also inhibit sucrase via noncompetitive inhibition. For instance, codeine and scopolamine inhibit the activity of yeast sucrase while castanospermine inhibits the activity of sucrase in human enterocyte-like cell line Caco-2Citation8,Citation9,Citation14.
The fluorescence measurements showed that there were structural changes after binding of the enzyme to the substrate (inhibitor). Binding of the enzyme to the substrate decreased the emission intensity of peak. Binding of ranitidine to pure enzyme or enzyme–substrate complex not only decreased the peak intensity but also revealed a red shift. This suggests that ranitidine could bind to both free enzyme and enzyme–substrate complex. Decreasing the peak intensity and the red-shift production suggest that a quenching has been occurred in free enzyme and enzyme–substrate complex after binding of ranitidine. This quenching proposed that the tryptophan residues displace to more polar medium, which may follow conformational changes in the enzyme. A recent report with fluorescence spectroscopy also indicated the structural changes in sucrase after binding of scopolamineCitation14.
Previous report indicated that scopolamine and codeine bind to sucrase at the site (inhibition site) rather than the active siteCitation14. Ranitidine may also bind to sucrase in the same manner of scopolamine and codeine, at the inhibition site. Considering three drugs (ranitidine, scopolamine, and codeine), which inhibit the sucrase with noncompetitive inhibition, a heterocyclic ring containing oxygen can be seen in their chemical structure. It seems that the oxygen of heterocyclic ring may have an important role in the binding of these drugs to the enzyme that is associated with conformational changes in the enzyme. However, the different groups attached to the heterocyclic ring affect the affinity of drugs in binding to the enzyme. Cimetidine with similar structure and effect to ranitidine has no affinity to bind to sucrase. Comparison between chemical structures of cimetidine and ranitidine showed that there is no oxygen bound heterocyclic ring in cimetidine molecule.
In conclusion, our results suggest that ranitidine binds to both free enzyme and enzyme–substrate complex and inhibit the enzyme by noncompetitive pattern, which is accompanied by conformational changes in the enzyme. Ranitidine has a heterocyclic ring containing oxygen that may play an important role in reaction between this inhibitor and the enzyme. The study of the effect of ranitidine on the sucrase–isomaltase enzyme of human or other mammalian gastrointestinal tract may reveal some similarities or difference between yeast sucrase and mammalian sucrase–isomaltase enzyme.
Declaration of interest
The authors declare no conflicts of interest.
References
- Pratt WB, Taylor P. Principles of drug action: The basis of pharmacology, 3rd Ed. Churchill Livingstone Inc., USA; 1990. pp. 396–397.
- Clark WG, Brater DC, Johnson AR. GOTH’S Medical pharmacology, 12th Ed. Mosby Company, USA; 1988. pp. 324–326.
- Gill M, Sanyal SN, Sareen ML. Effect of histamine H2-receptor antagonist, ranitidine on renal brush border and basolateral membranes. Res Exp Med (Berl) 1990;190:345–356.
- Vetvik K, Schrumpf E, Andersen KJ, Skagen DW, Halvorsen OJ. Effect of ranitidine on gastric and duodenal mucosal enzyme activities in duodenal ulcer patients. Scand J Gastroenterol 1990;25:1123–1128.
- Treem WR, McAdams L, Stanford L, Kastoff G, Justinich C, Hyams J. Sacrosidase therapy for congenital sucrase-isomaltase deficiency. J Pediatr Gastroenterol Nutr 1999;28:137–142.
- Treem WR, Ahsan N, Sullivan B, Rossi T, Holmes R, Fitzgerald J et al. Evaluation of liquid yeast-derived sucrase enzyme replacement in patients with sucrase-isomaltase deficiency. Gastroenterology 1993;105:1061–1068.
- Hanozet G, Pircher HP, Vanni P, Oesch B, Semenza G. An example of enzyme hysteresis. The slow and tight interaction of some fully competitive inhibitors with small intestinal sucrase. J Biol Chem 1981;256:3703–3711.
- Trugnan G, Rousset M, Zweibaum A. Castanospermine: a potent inhibitor of sucrase from the human enterocyte-like cell line Caco-2. FEBS Lett 1986;195:28–32.
- Minai-Tehrani D, Minoui S, Sepehre M, Sharif-Khodai Z, Aavani T. Inhibitory effect of codeine on sucrase activity. Drug Metab Lett 2009;3:58–60.
- Gascon S, Neumann NP, Lampen JO. Comparative study of the properties of the purified internal and external invertases from yeast. J Biol Chem 1968;243:1573–1577.
- Wu HL, Huang CH, Chen SH, Wu SM. Micellar electrokinetic chromatography of scopolamine-related anticholinergics. J Chromatogr A 1998;802:107–113.
- Robyt JF, White BJ. Biochemical techniques: Theory and practice. Brooks/Cole Pub. Com. Montery, California, USA; 1987. pp. 219–220.
- Yanase H, Iwata M, Kita K, Kato N, Tonomura K. Purification, crystallization and characterization of extracellular invertase from Zymomonas mobilis. J Ferment Bioeng 1995;79:367–369.
- Minai-Tehrani D, Fooladi N, Minoui S, Sobhani-Damavandifar Z, Aavani T, Heydarzadeh S et al. Structural changes and inhibition of sucrase after binding of scopolamine. Eur J Pharmacol 2010;635:23–26.
- Farooq N, Priyamvada S, Khan F, Yusufi AN. Time dependent effect of gentamicin on enzymes of carbohydrate metabolism and terminal digestion in rat intestine. Hum Exp Toxicol 2007;26:587–593.
- Chopra S, Saini RK, Sanyal SN. Intestinal toxicity of non-steroidal anti-inflammatory drugs with differential cyclooxygenase inhibition selectivity. Nutr Hosp 2007;22:528–537.
- Sim L, Willemsma C, Mohan S, Naim HY, Pinto BM, Rose DR. Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains. J Biol Chem 2010;285:17763–17770.